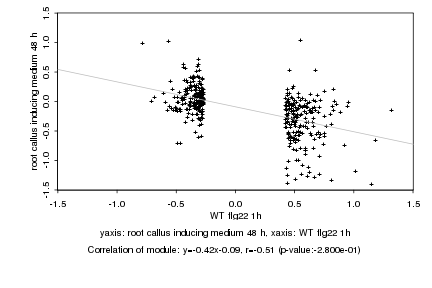
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT flg22 1h |
Log2 signal ratio
root callus inducing medium 48 h |
| 1 |
253503_at |
AT4G31950
|
CYP82C3, cytochrome P450, family 82, subfamily C, polypeptide 3 |
1.314 |
-0.150 |
| 2 |
248090_at |
AT5G55090
|
MAPKKK15, mitogen-activated protein kinase kinase kinase 15 |
1.182 |
-0.652 |
| 3 |
255945_at |
AT5G28610
|
unknown |
1.147 |
-1.391 |
| 4 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
1.007 |
-1.176 |
| 5 |
254249_at |
AT4G23280
|
CRK20, cysteine-rich RLK (RECEPTOR-like protein kinase) 20 |
0.952 |
-0.013 |
| 6 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
0.944 |
-0.070 |
| 7 |
249896_at |
AT5G22530
|
unknown |
0.913 |
-0.745 |
| 8 |
257846_at |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
0.886 |
-0.178 |
| 9 |
257511_at |
AT1G43000
|
[PLATZ transcription factor family protein] |
0.850 |
-0.048 |
| 10 |
254324_at |
AT4G22640
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.836 |
0.011 |
| 11 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.827 |
-0.143 |
| 12 |
266737_at |
AT2G47140
|
AtSDR5, SDR5, short-chain dehydrogenase reductase 5 |
0.820 |
0.209 |
| 13 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
0.815 |
-0.076 |
| 14 |
261394_at |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
0.810 |
-0.389 |
| 15 |
264202_at |
AT1G22810
|
[Integrase-type DNA-binding superfamily protein] |
0.803 |
-1.330 |
| 16 |
249618_at |
AT5G37490
|
[ARM repeat superfamily protein] |
0.795 |
-0.211 |
| 17 |
248775_at |
AT5G47850
|
CCR4, CRINKLY4 related 4 |
0.765 |
-0.408 |
| 18 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
0.750 |
-0.589 |
| 19 |
252944_at |
AT4G39320
|
[microtubule-associated protein-related] |
0.748 |
0.176 |
| 20 |
256968_at |
AT3G21070
|
ATNADK-1, NAD KINASE 1, NADK1, NAD kinase 1 |
0.747 |
-0.240 |
| 21 |
258364_at |
AT3G14225
|
GLIP4, GDSL-motif lipase 4 |
0.744 |
-0.535 |
| 22 |
247940_at |
AT5G57190
|
PSD2, phosphatidylserine decarboxylase 2 |
0.741 |
-0.714 |
| 23 |
254292_at |
AT4G23030
|
[MATE efflux family protein] |
0.716 |
0.024 |
| 24 |
250796_at |
AT5G05300
|
unknown |
0.714 |
-0.548 |
| 25 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.708 |
-0.605 |
| 26 |
254897_at |
AT4G11470
|
CRK31, cysteine-rich RLK (RECEPTOR-like protein kinase) 31 |
0.703 |
-0.092 |
| 27 |
260005_at |
AT1G67920
|
unknown |
0.703 |
-0.925 |
| 28 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
0.703 |
-1.225 |
| 29 |
248123_at |
AT5G54720
|
[Ankyrin repeat family protein] |
0.696 |
-0.186 |
| 30 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.695 |
-0.509 |
| 31 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
0.686 |
0.114 |
| 32 |
249001_at |
AT5G44990
|
[Glutathione S-transferase family protein] |
0.684 |
-0.554 |
| 33 |
261836_at |
AT1G16090
|
WAKL7, wall associated kinase-like 7 |
0.674 |
0.007 |
| 34 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.673 |
0.533 |
| 35 |
252873_at |
AT4G40020
|
[Myosin heavy chain-related protein] |
0.669 |
-0.138 |
| 36 |
254905_at |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
0.667 |
-0.088 |
| 37 |
247213_at |
AT5G64900
|
ATPEP1, ARABIDOPSIS THALIANA PEPTIDE 1, PEP1, PEPTIDE 1, PROPEP1, precursor of peptide 1 |
0.665 |
-0.642 |
| 38 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.661 |
-1.278 |
| 39 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
0.658 |
0.032 |
| 40 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.656 |
-0.419 |
| 41 |
250149_at |
AT5G14700
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.652 |
-0.784 |
| 42 |
249558_at |
AT5G38310
|
unknown |
0.651 |
-0.279 |
| 43 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
0.647 |
-0.109 |
| 44 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
0.646 |
-0.351 |
| 45 |
260394_at |
AT1G74080
|
ATMYB122, MYB DOMAIN PROTEIN 122, MYB122, myb domain protein 122 |
0.644 |
-0.171 |
| 46 |
254229_at |
AT4G23610
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.641 |
-0.186 |
| 47 |
257918_at |
AT3G23230
|
AtERF98, AtTDR1, ERF98, Ethylene Response Factor 98, TDR1, Transcriptional Regulator of Defense Response 1 |
0.638 |
-0.575 |
| 48 |
266052_at |
AT2G40740
|
ATWRKY55, WRKY DNA-BINDING PROTEIN 55, WRKY55, WRKY DNA-binding protein 55 |
0.631 |
-0.150 |
| 49 |
252346_at |
AT3G48650
|
[pseudogene] |
0.628 |
-0.171 |
| 50 |
259805_at |
AT1G47890
|
AtRLP7, receptor like protein 7, RLP7, receptor like protein 7 |
0.626 |
0.132 |
| 51 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.626 |
-0.534 |
| 52 |
252217_at |
AT3G50140
|
unknown |
0.624 |
-0.016 |
| 53 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.622 |
-0.137 |
| 54 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.618 |
-1.200 |
| 55 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.616 |
-1.110 |
| 56 |
248971_at |
AT5G45000
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.609 |
-0.354 |
| 57 |
264867_at |
AT1G24150
|
ATFH4, FORMIN HOMOLOGUE 4, FH4, formin homologue 4 |
0.605 |
-0.098 |
| 58 |
251950_at |
AT3G53600
|
[C2H2-type zinc finger family protein] |
0.605 |
-1.270 |
| 59 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.603 |
-0.022 |
| 60 |
253796_at |
AT4G28460
|
unknown |
0.602 |
-0.446 |
| 61 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
0.598 |
-0.195 |
| 62 |
257950_at |
AT3G21780
|
UGT71B6, UDP-glucosyl transferase 71B6 |
0.597 |
-0.085 |
| 63 |
263378_at |
AT2G40180
|
ATHPP2C5, phosphatase 2C5, PP2C5, phosphatase 2C5 |
0.597 |
-0.079 |
| 64 |
245082_at |
AT2G23270
|
unknown |
0.593 |
-0.354 |
| 65 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
0.590 |
-0.815 |
| 66 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.590 |
-0.826 |
| 67 |
254408_at |
AT4G21390
|
B120 |
0.587 |
-0.410 |
| 68 |
258752_at |
AT3G09520
|
ATEXO70H4, exocyst subunit exo70 family protein H4, EXO70H4, exocyst subunit exo70 family protein H4 |
0.586 |
-0.281 |
| 69 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
0.585 |
-0.789 |
| 70 |
265705_at |
AT2G03410
|
[Mo25 family protein] |
0.584 |
0.028 |
| 71 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.584 |
-0.897 |
| 72 |
256103_at |
AT1G13540
|
unknown |
0.583 |
-0.056 |
| 73 |
254014_at |
AT4G26120
|
[Ankyrin repeat family protein / BTB/POZ domain-containing protein] |
0.577 |
-0.161 |
| 74 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.575 |
-0.077 |
| 75 |
265643_at |
AT2G27390
|
[proline-rich family protein] |
0.574 |
-0.423 |
| 76 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.563 |
-1.070 |
| 77 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.562 |
0.059 |
| 78 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.561 |
-0.097 |
| 79 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
0.560 |
-0.526 |
| 80 |
253614_at |
AT4G30350
|
SMXL2, SMAX1-like 2 |
0.554 |
-0.216 |
| 81 |
259251_at |
AT3G07600
|
[Heavy metal transport/detoxification superfamily protein ] |
0.553 |
-1.232 |
| 82 |
246406_at |
AT1G57650
|
[ATP binding] |
0.549 |
-0.084 |
| 83 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.548 |
-0.232 |
| 84 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.548 |
-0.391 |
| 85 |
248814_at |
AT5G46910
|
[Transcription factor jumonji (jmj) family protein / zinc finger (C5HC2 type) family protein] |
0.546 |
-0.518 |
| 86 |
249225_at |
AT5G42140
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
0.545 |
0.026 |
| 87 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.544 |
-0.792 |
| 88 |
253044_at |
AT4G37290
|
unknown |
0.543 |
1.037 |
| 89 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.538 |
-0.568 |
| 90 |
249903_at |
AT5G22690
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.536 |
-0.428 |
| 91 |
245363_at |
AT4G15120
|
[VQ motif-containing protein] |
0.533 |
-0.062 |
| 92 |
251096_at |
AT5G01550
|
LecRK-VI.3, L-type lectin receptor kinase VI.3, LECRKA4.2, lectin receptor kinase a4.1 |
0.530 |
-0.106 |
| 93 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.529 |
-0.566 |
| 94 |
257583_at |
AT1G66480
|
[plastid movement impaired 2 (PMI2)] |
0.528 |
0.073 |
| 95 |
251769_at |
AT3G55950
|
ATCRR3, CCR3, CRINKLY4 related 3 |
0.528 |
-0.497 |
| 96 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.527 |
-0.989 |
| 97 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.526 |
0.146 |
| 98 |
264716_at |
AT1G70170
|
MMP, matrix metalloproteinase |
0.525 |
-0.651 |
| 99 |
266780_at |
AT2G29110
|
ATGLR2.8, glutamate receptor 2.8, GLR2.8, GLR2.8, glutamate receptor 2.8 |
0.521 |
-0.256 |
| 100 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.517 |
-0.216 |
| 101 |
248129_at |
AT5G54780
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.517 |
-0.120 |
| 102 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.516 |
-0.259 |
| 103 |
245089_at |
AT2G45290
|
TKL2, transketolase 2 |
0.515 |
-0.008 |
| 104 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
0.510 |
-0.245 |
| 105 |
251097_at |
AT5G01560
|
LecRK-VI.4, L-type lectin receptor kinase VI.4, LECRKA4.3, lectin receptor kinase a4.3 |
0.510 |
-0.256 |
| 106 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.510 |
-0.633 |
| 107 |
264314_at |
AT1G70420
|
unknown |
0.509 |
-0.987 |
| 108 |
249339_at |
AT5G41100
|
unknown |
0.509 |
-0.305 |
| 109 |
248708_at |
AT5G48560
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.509 |
-0.152 |
| 110 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.509 |
-0.510 |
| 111 |
254289_at |
AT4G22980
|
unknown |
0.508 |
-0.270 |
| 112 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
0.508 |
-0.826 |
| 113 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.506 |
-0.339 |
| 114 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.506 |
-1.307 |
| 115 |
261481_at |
AT1G14260
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.504 |
-0.244 |
| 116 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.503 |
-0.567 |
| 117 |
250575_at |
AT5G08240
|
unknown |
0.502 |
-0.421 |
| 118 |
260255_at |
AT1G74330
|
[Protein kinase superfamily protein] |
0.501 |
-0.098 |
| 119 |
262228_at |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
0.500 |
-0.682 |
| 120 |
267418_at |
AT2G35000
|
ATL9, Arabidopsis toxicos en levadura 9 |
0.496 |
-0.260 |
| 121 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
0.496 |
-0.616 |
| 122 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.494 |
-0.385 |
| 123 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
0.491 |
0.020 |
| 124 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
0.490 |
-0.199 |
| 125 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.490 |
-0.811 |
| 126 |
264660_at |
AT1G09940
|
HEMA2 |
0.488 |
-0.510 |
| 127 |
260439_at |
AT1G68340
|
unknown |
0.488 |
-0.048 |
| 128 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
0.488 |
-0.216 |
| 129 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.487 |
-0.655 |
| 130 |
262350_at |
AT2G48150
|
ATGPX4, glutathione peroxidase 4, GPX4, glutathione peroxidase 4 |
0.487 |
-0.156 |
| 131 |
249260_at |
AT5G41680
|
[Protein kinase superfamily protein] |
0.486 |
-0.026 |
| 132 |
265620_at |
AT2G27310
|
[F-box family protein] |
0.485 |
-0.626 |
| 133 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.484 |
-0.136 |
| 134 |
252345_at |
AT3G48640
|
unknown |
0.484 |
-0.061 |
| 135 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
0.482 |
-0.302 |
| 136 |
265024_at |
AT1G24600
|
unknown |
0.482 |
0.255 |
| 137 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
0.482 |
-0.327 |
| 138 |
260754_at |
AT1G49000
|
unknown |
0.481 |
0.225 |
| 139 |
254857_at |
AT4G12120
|
ATSEC1B, SEC1B |
0.480 |
-0.322 |
| 140 |
254469_at |
AT4G20470
|
unknown |
0.478 |
0.060 |
| 141 |
255999_at |
AT1G29860
|
ATWRKY71, WRKY DNA-BINDING PROTEIN 71, WRKY71, WRKY DNA-binding protein 71 |
0.474 |
-0.055 |
| 142 |
257416_at |
AT2G17750
|
NIP1, NEP-interacting protein 1 |
0.473 |
-0.042 |
| 143 |
247240_at |
AT5G64660
|
ATCMPG2, CMPG2, CYS, MET, PRO, and GLY protein 2 |
0.472 |
-0.064 |
| 144 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.467 |
-0.387 |
| 145 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.467 |
-0.112 |
| 146 |
260557_at |
AT2G43610
|
[Chitinase family protein] |
0.467 |
-0.277 |
| 147 |
251379_at |
AT3G60680
|
unknown |
0.466 |
0.082 |
| 148 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.466 |
-0.164 |
| 149 |
245662_at |
AT1G28190
|
unknown |
0.466 |
-0.603 |
| 150 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
0.464 |
-0.439 |
| 151 |
247047_at |
AT5G66650
|
unknown |
0.463 |
-0.660 |
| 152 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.461 |
-0.584 |
| 153 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.459 |
-0.704 |
| 154 |
245156_at |
AT5G12480
|
CPK7, calmodulin-domain protein kinase 7 |
0.456 |
0.147 |
| 155 |
260040_at |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
0.455 |
0.537 |
| 156 |
256428_at |
AT3G11080
|
AtRLP35, receptor like protein 35, RLP35, receptor like protein 35 |
0.455 |
-0.025 |
| 157 |
254647_at |
AT4G18540
|
unknown |
0.455 |
-0.017 |
| 158 |
251832_at |
AT3G55150
|
ATEXO70H1, exocyst subunit exo70 family protein H1, EXO70H1, exocyst subunit exo70 family protein H1 |
0.454 |
-0.594 |
| 159 |
248719_at |
AT5G47910
|
ATRBOHD, RBOHD, respiratory burst oxidase homologue D |
0.454 |
-0.385 |
| 160 |
248706_at |
AT5G48530
|
unknown |
0.452 |
-0.752 |
| 161 |
255342_at |
AT4G04510
|
CRK38, cysteine-rich RLK (RECEPTOR-like protein kinase) 38 |
0.452 |
-0.112 |
| 162 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
0.451 |
-0.121 |
| 163 |
248176_at |
AT5G54650
|
ATFH5, FORMIN HOMOLOGY 5, Fh5, formin homology5 |
0.450 |
-0.057 |
| 164 |
246370_at |
AT1G51920
|
unknown |
0.448 |
-0.551 |
| 165 |
249314_at |
AT5G41180
|
[leucine-rich repeat transmembrane protein kinase family protein] |
0.448 |
0.039 |
| 166 |
250920_at |
AT5G03390
|
unknown |
0.445 |
-0.524 |
| 167 |
267576_at |
AT2G30640
|
MUG2, MUSTANG 2 |
0.445 |
-0.057 |
| 168 |
258282_at |
AT3G26910
|
[hydroxyproline-rich glycoprotein family protein] |
0.444 |
-0.287 |
| 169 |
253827_at |
AT4G28085
|
unknown |
0.442 |
-1.008 |
| 170 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.441 |
0.014 |
| 171 |
257277_at |
AT3G14470
|
[NB-ARC domain-containing disease resistance protein] |
0.441 |
-0.190 |
| 172 |
254487_at |
AT4G20780
|
CML42, calmodulin like 42 |
0.440 |
-0.619 |
| 173 |
252825_at |
AT4G39890
|
AtRABH1c, RAB GTPase homolog H1C, RABH1c, RAB GTPase homolog H1C |
0.440 |
-0.527 |
| 174 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
0.439 |
-1.239 |
| 175 |
251277_at |
AT3G61760
|
ADL1B, DYNAMIN-like 1B, DL1B, DYNAMIN-like 1B |
0.438 |
0.137 |
| 176 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.437 |
-0.227 |
| 177 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
0.437 |
-0.355 |
| 178 |
262360_at |
AT1G73080
|
ATPEPR1, PEP1 RECEPTOR 1, PEPR1, PEP1 receptor 1 |
0.436 |
-0.329 |
| 179 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.436 |
-1.389 |
| 180 |
260434_at |
AT1G68330
|
unknown |
0.435 |
0.104 |
| 181 |
245566_at |
AT4G14610
|
[pseudogene] |
0.435 |
-0.344 |
| 182 |
265030_at |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
0.435 |
0.211 |
| 183 |
263108_at |
AT1G65240
|
[Eukaryotic aspartyl protease family protein] |
0.433 |
0.004 |
| 184 |
247137_at |
AT5G66210
|
CPK28, calcium-dependent protein kinase 28 |
0.431 |
-0.255 |
| 185 |
256583_at |
AT3G28850
|
[Glutaredoxin family protein] |
0.431 |
-0.059 |
| 186 |
263182_at |
AT1G05575
|
unknown |
0.429 |
-1.123 |
| 187 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.426 |
-0.144 |
| 188 |
253063_at |
AT4G37640
|
ACA2, calcium ATPase 2 |
0.426 |
-0.060 |
| 189 |
248091_at |
AT5G55120
|
galactose-1-phosphate guanylyltransferase (GDP)s;GDP-D-glucose phosphorylases;quercetin 4'-O-glucosyltransferases |
0.426 |
-0.006 |
| 190 |
251061_at |
AT5G01830
|
SAUR21, SMALL AUXIN UP RNA 21 |
0.425 |
-0.132 |
| 191 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.424 |
-0.437 |
| 192 |
260362_at |
AT1G70530
|
CRK3, cysteine-rich RLK (RECEPTOR-like protein kinase) 3 |
0.423 |
-0.208 |
| 193 |
259734_at |
AT1G77500
|
unknown |
0.423 |
-0.074 |
| 194 |
265728_at |
AT2G31990
|
[Exostosin family protein] |
0.422 |
-0.359 |
| 195 |
253665_at |
AT4G30230
|
unknown |
0.421 |
-0.125 |
| 196 |
258577_at |
AT3G04220
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.420 |
-0.436 |
| 197 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.420 |
-0.064 |
| 198 |
246532_at |
AT5G15870
|
[glycosyl hydrolase family 81 protein] |
0.419 |
-0.297 |
| 199 |
254575_at |
AT4G19460
|
[UDP-Glycosyltransferase superfamily protein] |
0.419 |
0.061 |
| 200 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.789 |
0.997 |
| 201 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.714 |
0.007 |
| 202 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
-0.687 |
0.072 |
| 203 |
246929_at |
AT5G25210
|
unknown |
-0.609 |
0.141 |
| 204 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.593 |
-0.005 |
| 205 |
261817_at |
AT1G08180
|
unknown |
-0.581 |
-0.149 |
| 206 |
256789_at |
AT3G13672
|
SINA2 |
-0.567 |
1.029 |
| 207 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.563 |
-0.070 |
| 208 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.555 |
0.354 |
| 209 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.542 |
-0.115 |
| 210 |
265880_at |
AT2G42300
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.540 |
0.210 |
| 211 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
-0.531 |
-0.136 |
| 212 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.522 |
0.084 |
| 213 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
-0.514 |
-0.139 |
| 214 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
-0.512 |
-0.090 |
| 215 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
-0.503 |
-0.114 |
| 216 |
259302_at |
AT3G05120
|
ATGID1A, GA INSENSITIVE DWARF1A, GID1A, GA INSENSITIVE DWARF1A |
-0.494 |
-0.163 |
| 217 |
247151_at |
AT5G65640
|
bHLH093, beta HLH protein 93 |
-0.493 |
0.079 |
| 218 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.492 |
-0.701 |
| 219 |
249532_at |
AT5G38780
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.492 |
-0.004 |
| 220 |
260799_at |
AT1G78270
|
AtUGT85A4, UDP-glucosyl transferase 85A4, UGT85A4, UDP-glucosyl transferase 85A4 |
-0.488 |
-0.015 |
| 221 |
267637_at |
AT2G42190
|
unknown |
-0.480 |
0.157 |
| 222 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.477 |
-0.123 |
| 223 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.470 |
-0.700 |
| 224 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.469 |
-0.155 |
| 225 |
256455_at |
AT1G75190
|
unknown |
-0.466 |
-0.130 |
| 226 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
-0.459 |
0.063 |
| 227 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
-0.452 |
0.072 |
| 228 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.451 |
-0.035 |
| 229 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.448 |
0.025 |
| 230 |
259122_at |
AT3G02210
|
COBL1, COBRA-like protein 1 precursor |
-0.443 |
0.578 |
| 231 |
246576_at |
AT1G31650
|
ATROPGEF14, ROPGEF14, ROP (rho of plants) guanine nucleotide exchange factor 14 |
-0.441 |
0.636 |
| 232 |
261607_at |
AT1G49660
|
AtCXE5, carboxyesterase 5, CXE5, carboxyesterase 5 |
-0.439 |
0.049 |
| 233 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.437 |
0.118 |
| 234 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
-0.436 |
0.069 |
| 235 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
-0.435 |
0.358 |
| 236 |
259240_at |
AT3G11590
|
unknown |
-0.433 |
0.045 |
| 237 |
261609_at |
AT1G49740
|
[PLC-like phosphodiesterases superfamily protein] |
-0.429 |
0.572 |
| 238 |
257879_at |
AT3G17160
|
unknown |
-0.428 |
0.182 |
| 239 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
-0.428 |
-0.345 |
| 240 |
267462_at |
AT2G33735
|
[Chaperone DnaJ-domain superfamily protein] |
-0.427 |
0.204 |
| 241 |
253317_at |
AT4G33960
|
unknown |
-0.426 |
-0.032 |
| 242 |
254853_at |
AT4G12080
|
AHL1, AT-hook motif nuclear-localized protein 1, ATAHL1 |
-0.418 |
-0.119 |
| 243 |
252184_at |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
-0.418 |
0.372 |
| 244 |
248820_at |
AT5G47060
|
unknown |
-0.415 |
0.128 |
| 245 |
252272_at |
AT3G49670
|
BAM2, BARELY ANY MERISTEM 2 |
-0.413 |
0.326 |
| 246 |
262811_at |
AT1G11700
|
unknown |
-0.413 |
-0.126 |
| 247 |
264930_at |
AT1G60800
|
AtNIK3, NIK3, NSP-interacting kinase 3 |
-0.407 |
0.148 |
| 248 |
247716_at |
AT5G59350
|
unknown |
-0.407 |
0.235 |
| 249 |
254011_at |
AT4G26370
|
[antitermination NusB domain-containing protein] |
-0.406 |
0.193 |
| 250 |
246997_at |
AT5G67390
|
unknown |
-0.405 |
-0.269 |
| 251 |
258663_at |
AT3G08670
|
unknown |
-0.403 |
-0.194 |
| 252 |
256784_at |
AT3G13674
|
unknown |
-0.396 |
0.228 |
| 253 |
245830_at |
AT1G57790
|
[F-box family protein] |
-0.396 |
-0.052 |
| 254 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
-0.392 |
-0.129 |
| 255 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.390 |
-0.048 |
| 256 |
266929_at |
AT2G45850
|
AHL9, AT-hook motif nuclear localized protein 9 |
-0.390 |
-0.127 |
| 257 |
267606_at |
AT2G26640
|
KCS11, 3-ketoacyl-CoA synthase 11 |
-0.388 |
0.013 |
| 258 |
258424_at |
AT3G16750
|
unknown |
-0.387 |
-0.069 |
| 259 |
246393_at |
AT1G58150
|
unknown |
-0.386 |
0.058 |
| 260 |
251936_at |
AT3G53700
|
MEE40, maternal effect embryo arrest 40 |
-0.386 |
0.336 |
| 261 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.384 |
-0.002 |
| 262 |
267048_at |
AT2G34200
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.384 |
0.245 |
| 263 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
-0.382 |
0.284 |
| 264 |
249612_at |
AT5G37290
|
[ARM repeat superfamily protein] |
-0.380 |
0.246 |
| 265 |
256741_at |
AT3G29375
|
[XH domain-containing protein] |
-0.380 |
0.416 |
| 266 |
246518_at |
AT5G15770
|
AtGNA1, glucose-6-phosphate acetyltransferase 1, GNA1, glucose-6-phosphate acetyltransferase 1 |
-0.376 |
0.199 |
| 267 |
263737_at |
AT1G60010
|
unknown |
-0.373 |
0.271 |
| 268 |
246144_at |
AT5G20110
|
[Dynein light chain type 1 family protein] |
-0.369 |
-0.218 |
| 269 |
246439_at |
AT5G17600
|
[RING/U-box superfamily protein] |
-0.368 |
0.066 |
| 270 |
258332_at |
AT3G16180
|
NRT1.12, nitrate transporter 1.12 |
-0.366 |
-0.310 |
| 271 |
253692_at |
AT4G29720
|
ATPAO5, polyamine oxidase 5, PAO5, polyamine oxidase 5 |
-0.365 |
0.423 |
| 272 |
253475_at |
AT4G32290
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.363 |
0.399 |
| 273 |
253270_at |
AT4G34160
|
CYCD3, CYCD3;1, CYCLIN D3;1 |
-0.362 |
0.430 |
| 274 |
245592_at |
AT4G14540
|
NF-YB3, nuclear factor Y, subunit B3 |
-0.361 |
-0.058 |
| 275 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.361 |
-0.033 |
| 276 |
249763_at |
AT5G24010
|
[Protein kinase superfamily protein] |
-0.358 |
0.393 |
| 277 |
245644_at |
AT1G25320
|
[Leucine-rich repeat protein kinase family protein] |
-0.356 |
0.342 |
| 278 |
254810_at |
AT4G12390
|
PME1, pectin methylesterase inhibitor 1 |
-0.354 |
0.269 |
| 279 |
248773_at |
AT5G47820
|
FRA1, FRAGILE FIBER 1 |
-0.354 |
0.056 |
| 280 |
250126_at |
AT5G16310
|
UCH1 |
-0.353 |
0.109 |
| 281 |
253811_at |
AT4G28190
|
ULT, ULTRAPETALA, ULT1, ULTRAPETALA1 |
-0.353 |
-0.011 |
| 282 |
260474_at |
AT1G11090
|
[alpha/beta-Hydrolases superfamily protein] |
-0.352 |
0.094 |
| 283 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
-0.352 |
-0.027 |
| 284 |
265718_at |
AT2G03340
|
WRKY3, WRKY DNA-binding protein 3 |
-0.351 |
-0.083 |
| 285 |
253854_at |
AT4G27900
|
[CCT motif family protein] |
-0.350 |
0.113 |
| 286 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
-0.349 |
0.409 |
| 287 |
262668_at |
AT1G62830
|
ATLSD1, ARABIDOPSIS LYSINE-SPECIFIC HISTONE DEMETHYLASE, ATSWP1, LDL1, LSD1-like 1, LSD1, LYSINE-SPECIFIC HISTONE DEMETHYLASE, SWP1 |
-0.349 |
0.284 |
| 288 |
267135_at |
AT2G23430
|
ICK1, KRP1, KIP-RELATED PROTEIN 1 |
-0.349 |
-0.067 |
| 289 |
261048_at |
AT1G01420
|
UGT72B3, UDP-glucosyl transferase 72B3 |
-0.347 |
-0.111 |
| 290 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
-0.346 |
-0.076 |
| 291 |
261559_at |
AT1G01780
|
PLIM2b, PLIM2b |
-0.344 |
-0.512 |
| 292 |
252185_at |
AT3G50780
|
unknown |
-0.340 |
0.210 |
| 293 |
262631_at |
AT1G06500
|
unknown |
-0.339 |
0.202 |
| 294 |
251008_at |
AT5G02710
|
unknown |
-0.338 |
0.126 |
| 295 |
261552_at |
AT1G63430
|
[Leucine-rich repeat protein kinase family protein] |
-0.338 |
-0.086 |
| 296 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
-0.337 |
0.028 |
| 297 |
254427_at |
AT4G21190
|
emb1417, embryo defective 1417 |
-0.336 |
0.084 |
| 298 |
245861_at |
AT5G28300
|
AtGT2L, GT2L, GT-2Like protein |
-0.336 |
-0.132 |
| 299 |
255224_at |
AT4G05400
|
[copper ion binding] |
-0.335 |
0.272 |
| 300 |
248423_at |
AT5G51670
|
unknown |
-0.335 |
0.517 |
| 301 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.333 |
0.254 |
| 302 |
246901_at |
AT5G25630
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.331 |
-0.036 |
| 303 |
265967_at |
AT2G37450
|
UMAMIT13, Usually multiple acids move in and out Transporters 13 |
-0.331 |
-0.219 |
| 304 |
259863_at |
AT1G72630
|
ELF4-L2, ELF4-like 2 |
-0.330 |
0.147 |
| 305 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
-0.327 |
0.071 |
| 306 |
263438_at |
AT2G28660
|
[Chloroplast-targeted copper chaperone protein] |
-0.327 |
0.136 |
| 307 |
258209_at |
AT3G14060
|
unknown |
-0.327 |
0.606 |
| 308 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
-0.326 |
0.425 |
| 309 |
255428_at |
AT4G03390
|
SRF3, STRUBBELIG-receptor family 3 |
-0.325 |
0.072 |
| 310 |
265027_at |
AT1G24450
|
NFD2, NUCLEAR FUSION DEFECTIVE 2 |
-0.323 |
0.132 |
| 311 |
262952_at |
AT1G75770
|
unknown |
-0.323 |
-0.011 |
| 312 |
250156_at |
AT5G15170
|
TDP1, tyrosyl-DNA phosphodiesterase 1 |
-0.322 |
0.087 |
| 313 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.321 |
-0.206 |
| 314 |
247248_at |
AT5G64560
|
ATMGT9, MGT9, magnesium transporter 9, MRS2-2 |
-0.320 |
0.411 |
| 315 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
-0.318 |
0.092 |
| 316 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.318 |
-0.597 |
| 317 |
255551_at |
AT4G01840
|
ATKCO5, ATTPK5, KCO5, Ca2+ activated outward rectifying K+ channel 5, TPK5 |
-0.317 |
0.135 |
| 318 |
245901_at |
AT5G11060
|
KNAT4, KNOTTED1-like homeobox gene 4 |
-0.317 |
-0.076 |
| 319 |
259207_at |
AT3G09050
|
unknown |
-0.314 |
-0.105 |
| 320 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.314 |
-0.283 |
| 321 |
255128_at |
AT4G08310
|
unknown |
-0.314 |
0.133 |
| 322 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.314 |
-0.099 |
| 323 |
249118_at |
AT5G43870
|
[FUNCTIONS IN: phosphoinositide binding] |
-0.314 |
0.715 |
| 324 |
265663_at |
AT2G24290
|
unknown |
-0.314 |
-0.108 |
| 325 |
253054_at |
AT4G37580
|
COP3, CONSTITUTIVE PHOTOMORPHOGENIC 3, HLS1, HOOKLESS 1, UNS2, UNUSUAL SUGAR RESPONSE 2 |
-0.313 |
0.633 |
| 326 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.313 |
0.248 |
| 327 |
263688_at |
AT1G26920
|
unknown |
-0.313 |
0.015 |
| 328 |
262170_at |
AT1G74940
|
unknown |
-0.312 |
-0.292 |
| 329 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.312 |
-0.071 |
| 330 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
-0.312 |
-0.169 |
| 331 |
254803_at |
AT4G13100
|
[RING/U-box superfamily protein] |
-0.311 |
0.310 |
| 332 |
259300_at |
AT3G05100
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.310 |
-0.033 |
| 333 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.310 |
-0.140 |
| 334 |
261616_at |
AT1G33060
|
ANAC014, NAC 014, NAC014, NAC 014 |
-0.310 |
0.158 |
| 335 |
252224_at |
AT3G49860
|
ARLA1B, ADP-ribosylation factor-like A1B, ATARLA1B, ADP-ribosylation factor-like A1B |
-0.309 |
-0.398 |
| 336 |
255583_at |
AT4G01510
|
ARV2 |
-0.308 |
0.021 |
| 337 |
248046_at |
AT5G56040
|
SKM2, STERILITY-REGULATING KINASE MEMBER 2 |
-0.307 |
0.198 |
| 338 |
262071_at |
AT1G59510
|
CF9 |
-0.305 |
0.110 |
| 339 |
254834_at |
AT4G12300
|
CYP706A4, cytochrome P450, family 706, subfamily A, polypeptide 4 |
-0.305 |
0.355 |
| 340 |
255829_at |
AT2G40540
|
ATKT2, ATKUP2, KT2, potassium transporter 2, KUP2, SHY3, TRK2 |
-0.304 |
0.420 |
| 341 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
-0.304 |
0.538 |
| 342 |
260371_at |
AT1G69690
|
AtTCP15, TCP15, TEOSINTE BRANCHED1/CYCLOIDEA/PCF 15 |
-0.304 |
0.072 |
| 343 |
261112_at |
AT1G75420
|
[UDP-Glycosyltransferase superfamily protein] |
-0.303 |
0.160 |
| 344 |
259293_at |
AT3G11580
|
[AP2/B3-like transcriptional factor family protein] |
-0.303 |
-0.308 |
| 345 |
251929_at |
AT3G53920
|
SIG3, SIGMA FACTOR 3, SIGC, RNApolymerase sigma-subunit C |
-0.302 |
-0.022 |
| 346 |
264096_at |
AT1G78995
|
unknown |
-0.301 |
-0.015 |
| 347 |
259812_at |
AT1G49840
|
unknown |
-0.299 |
0.018 |
| 348 |
248669_at |
AT5G48730
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.298 |
0.298 |
| 349 |
250752_at |
AT5G05690
|
CBB3, CABBAGE 3, CPD, CONSTITUTIVE PHOTOMORPHOGENIC DWARF, CYP90, CYP90A, CYP90A1, CYTOCHROME P450 90A1, DWF3, DWARF 3 |
-0.296 |
-0.058 |
| 350 |
261658_at |
AT1G50040
|
unknown |
-0.296 |
-0.082 |
| 351 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.296 |
-0.040 |
| 352 |
257236_at |
AT3G15095
|
HCF243, high chlorophyll fluorescence 243 |
-0.296 |
0.111 |
| 353 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
-0.295 |
-0.259 |
| 354 |
264212_at |
AT1G22660
|
[Polynucleotide adenylyltransferase family protein] |
-0.295 |
0.234 |
| 355 |
265884_at |
AT2G42320
|
[nucleolar protein gar2-related] |
-0.295 |
-0.593 |
| 356 |
264510_at |
AT1G09530
|
PAP3, PHYTOCHROME-ASSOCIATED PROTEIN 3, PIF3, phytochrome interacting factor 3, POC1, PHOTOCURRENT 1 |
-0.294 |
-0.014 |
| 357 |
257451_at |
AT1G05690
|
BT3, BTB and TAZ domain protein 3 |
-0.293 |
0.004 |
| 358 |
255535_at |
AT4G01790
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
-0.293 |
0.221 |
| 359 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
-0.293 |
0.180 |
| 360 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
-0.291 |
0.086 |
| 361 |
258530_at |
AT3G06840
|
unknown |
-0.290 |
0.187 |
| 362 |
261951_at |
AT1G64490
|
[DEK, chromatin associated protein] |
-0.290 |
0.303 |
| 363 |
251500_at |
AT3G59110
|
[Protein kinase superfamily protein] |
-0.290 |
0.159 |
| 364 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
-0.290 |
-0.388 |
| 365 |
265869_at |
AT2G01760
|
ARR14, response regulator 14, RR14, response regulator 14 |
-0.289 |
-0.004 |
| 366 |
251326_at |
AT3G61590
|
HS, HWS, HAWAIIAN SKIRT |
-0.289 |
0.068 |
| 367 |
264057_at |
AT2G28550
|
RAP2.7, related to AP2.7, TOE1, TARGET OF EARLY ACTIVATION TAGGED (EAT) 1 |
-0.288 |
-0.171 |
| 368 |
259818_at |
AT1G49890
|
QWRF2, QWRF domain containing 2 |
-0.287 |
0.115 |
| 369 |
250462_at |
AT5G10020
|
SIRK1, sucrose-induced receptor kinase 1 |
-0.287 |
0.106 |
| 370 |
246231_at |
AT4G37080
|
unknown |
-0.287 |
0.052 |
| 371 |
261451_at |
AT1G21060
|
unknown |
-0.286 |
0.383 |
| 372 |
264700_at |
AT1G70100
|
unknown |
-0.285 |
-0.278 |
| 373 |
253243_at |
AT4G34560
|
unknown |
-0.285 |
0.225 |
| 374 |
267093_at |
AT2G38170
|
ATCAX1, CAX1, cation exchanger 1, RCI4, RARE COLD INDUCIBLE 4 |
-0.285 |
-0.272 |
| 375 |
262421_at |
AT1G50290
|
unknown |
-0.284 |
-0.157 |
| 376 |
263597_at |
AT2G01870
|
unknown |
-0.284 |
0.017 |
| 377 |
246782_at |
AT5G27320
|
ATGID1C, GID1C, GA INSENSITIVE DWARF1C |
-0.284 |
0.149 |
| 378 |
262727_at |
AT1G75800
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.283 |
-0.041 |
| 379 |
256331_at |
AT1G76880
|
[Duplicated homeodomain-like superfamily protein] |
-0.283 |
-0.160 |
| 380 |
264054_at |
AT2G22540
|
AGL22, AGAMOUS-like 22, FAQ1, Flowering Arabidopsis QTL1, SVP, SHORT VEGETATIVE PHASE |
-0.282 |
-0.012 |
| 381 |
255449_at |
AT4G02820
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.282 |
0.397 |
| 382 |
259617_at |
AT1G47970
|
unknown |
-0.282 |
0.125 |
| 383 |
246446_at |
AT5G17640
|
ASG1, Abiotic Stress Gene 1 |
-0.281 |
0.054 |
| 384 |
267508_at |
AT2G45700
|
[sterile alpha motif (SAM) domain-containing protein] |
-0.281 |
0.081 |
| 385 |
251302_at |
AT3G61970
|
NGA2, NGATHA2 |
-0.281 |
-0.123 |
| 386 |
250718_at |
AT5G06240
|
emb2735, embryo defective 2735 |
-0.281 |
0.071 |
| 387 |
263067_at |
AT2G17550
|
TRM26, TON1 Recruiting Motif 26 |
-0.279 |
-0.214 |
| 388 |
262230_at |
AT1G68560
|
ATXYL1, ALPHA-XYLOSIDASE 1, AXY3, altered xyloglucan 3, TRG1, thermoinhibition resistant germination 1, XYL1, alpha-xylosidase 1 |
-0.278 |
0.131 |
| 389 |
249999_at |
AT5G18640
|
[alpha/beta-Hydrolases superfamily protein] |
-0.278 |
-0.044 |
| 390 |
253253_at |
AT4G34750
|
SAUR49, SMALL AUXIN UPREGULATED RNA 49 |
-0.278 |
0.041 |
| 391 |
255436_at |
AT4G03150
|
unknown |
-0.277 |
0.048 |
| 392 |
253669_at |
AT4G30000
|
[Dihydropterin pyrophosphokinase / Dihydropteroate synthase] |
-0.276 |
0.162 |
| 393 |
254682_at |
AT4G13640
|
UNE16, unfertilized embryo sac 16 |
-0.276 |
0.055 |
| 394 |
257712_at |
AT3G27420
|
unknown |
-0.274 |
-0.032 |
| 395 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.274 |
-0.010 |
| 396 |
256516_at |
AT1G66150
|
TMK1, transmembrane kinase 1 |
-0.274 |
0.023 |
| 397 |
254218_at |
AT4G23740
|
[Leucine-rich repeat protein kinase family protein] |
-0.274 |
0.215 |