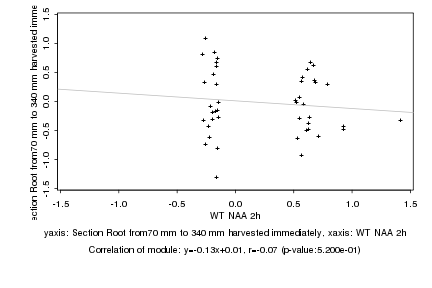
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT NAA 2h |
Log2 signal ratio
Section Root from70 mm to 340 mm harvested immediately |
| 1 |
245076_at |
AT2G23170
|
GH3.3 |
1.412 |
-0.319 |
| 2 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.921 |
-0.415 |
| 3 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.920 |
-0.475 |
| 4 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.783 |
0.300 |
| 5 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.705 |
-0.586 |
| 6 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.686 |
0.339 |
| 7 |
247474_at |
AT5G62280
|
unknown |
0.672 |
0.376 |
| 8 |
248279_at |
AT5G52910
|
ATIM, TIMELESS |
0.669 |
0.624 |
| 9 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
0.638 |
0.688 |
| 10 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
0.633 |
-0.272 |
| 11 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.625 |
-0.371 |
| 12 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.621 |
-0.468 |
| 13 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
0.610 |
0.558 |
| 14 |
248509_at |
AT5G50335
|
unknown |
0.602 |
-0.496 |
| 15 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
0.583 |
-0.037 |
| 16 |
248801_at |
AT5G47370
|
HAT2 |
0.570 |
0.421 |
| 17 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
0.562 |
0.355 |
| 18 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
0.558 |
-0.920 |
| 19 |
252204_at |
AT3G50340
|
unknown |
0.546 |
-0.290 |
| 20 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
0.543 |
0.079 |
| 21 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
0.528 |
-0.634 |
| 22 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
0.522 |
-0.002 |
| 23 |
262263_at |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
0.513 |
0.030 |
| 24 |
256784_at |
AT3G13674
|
unknown |
-0.289 |
0.814 |
| 25 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
-0.278 |
-0.310 |
| 26 |
262811_at |
AT1G11700
|
unknown |
-0.267 |
0.329 |
| 27 |
258343_at |
AT3G22810
|
[FUNCTIONS IN: phosphoinositide binding] |
-0.260 |
-0.735 |
| 28 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.259 |
1.098 |
| 29 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.233 |
-0.428 |
| 30 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
-0.223 |
-0.606 |
| 31 |
250158_at |
AT5G15190
|
unknown |
-0.221 |
-0.069 |
| 32 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
-0.203 |
-0.176 |
| 33 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.197 |
-0.309 |
| 34 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
-0.190 |
0.476 |
| 35 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
-0.185 |
0.861 |
| 36 |
262244_at |
AT1G48260
|
CIPK17, CBL-interacting protein kinase 17, SnRK3.21, SNF1-RELATED PROTEIN KINASE 3.21 |
-0.177 |
-0.155 |
| 37 |
251049_at |
AT5G02430
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.171 |
0.609 |
| 38 |
251725_at |
AT3G56260
|
unknown |
-0.168 |
0.672 |
| 39 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
-0.168 |
-1.301 |
| 40 |
245411_at |
AT4G17240
|
unknown |
-0.166 |
0.305 |
| 41 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.160 |
-0.803 |
| 42 |
260903_at |
AT1G02460
|
[Pectin lyase-like superfamily protein] |
-0.160 |
-0.144 |
| 43 |
253101_at |
AT4G37430
|
CYP81F1, CYTOCHROME P450 MONOOXYGENASE 81F1, CYP91A2, cytochrome P450, family 91, subfamily A, polypeptide 2 |
-0.159 |
0.750 |
| 44 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.148 |
-0.005 |
| 45 |
246900_at |
AT5G25620
|
AtYUC6, YUC6, YUCCA6 |
-0.146 |
-0.275 |