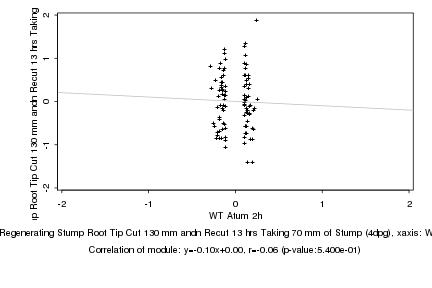
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT Atum 2h |
Log2 signal ratio
Regenerating Stump Root Tip Cut 130 mm andn Recut 13 hrs Taking 70 mm of Stump (4dpg) |
| 1 |
247216_at |
AT5G64860
|
DPE1, disproportionating enzyme |
0.252 |
0.062 |
| 2 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
0.233 |
1.897 |
| 3 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
0.212 |
-0.146 |
| 4 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
0.208 |
-0.632 |
| 5 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
0.200 |
-0.206 |
| 6 |
264314_at |
AT1G70420
|
unknown |
0.195 |
-0.868 |
| 7 |
249639_at |
AT5G36930
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.192 |
-0.613 |
| 8 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.188 |
-1.410 |
| 9 |
264874_at |
AT1G24240
|
[Ribosomal protein L19 family protein] |
0.168 |
-0.863 |
| 10 |
258006_at |
AT3G19400
|
[Cysteine proteinases superfamily protein] |
0.165 |
-0.085 |
| 11 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
0.160 |
-0.099 |
| 12 |
248848_at |
AT5G46520
|
VICTR, VARIATION IN COMPOUND TRIGGERED ROOT growth response |
0.158 |
-0.261 |
| 13 |
253581_at |
AT4G30660
|
[Low temperature and salt responsive protein family] |
0.156 |
0.400 |
| 14 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
0.156 |
-0.278 |
| 15 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
0.147 |
0.320 |
| 16 |
255743_at |
AT1G25375
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
0.147 |
0.555 |
| 17 |
248708_at |
AT5G48560
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.144 |
0.608 |
| 18 |
251853_at |
AT3G54790
|
[ARM repeat superfamily protein] |
0.142 |
0.132 |
| 19 |
255232_at |
AT4G05330
|
AGD13, ARF-GAP domain 13 |
0.137 |
-1.405 |
| 20 |
251641_at |
AT3G57470
|
[Insulinase (Peptidase family M16) family protein] |
0.135 |
-0.196 |
| 21 |
250419_at |
AT5G11250
|
[Disease resistance protein (TIR-NBS-LRR class)] |
0.132 |
-0.461 |
| 22 |
262768_at |
AT1G12990
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
0.132 |
-0.273 |
| 23 |
245254_at |
AT4G14680
|
APS3 |
0.127 |
0.113 |
| 24 |
245301_at |
AT4G17190
|
FPS2, farnesyl diphosphate synthase 2 |
0.124 |
0.502 |
| 25 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
0.123 |
-0.555 |
| 26 |
250810_at |
AT5G05090
|
[Homeodomain-like superfamily protein] |
0.123 |
-0.724 |
| 27 |
262666_at |
AT1G14080
|
ATFUT6, FUT6, fucosyltransferase 6 |
0.122 |
0.396 |
| 28 |
247313_at |
AT5G63980
|
ALX8, ALTERED EXPRESSION OF APX2 8, AtFRY1, ATSAL1, FRY1, FIERY1, HOS2, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 2, RON1, ROTUNDA 1, SAL1, SUPO1, suppressors of PIN1 overexpression 1 |
0.122 |
0.870 |
| 29 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
0.119 |
-0.235 |
| 30 |
245502_at |
AT4G15640
|
unknown |
0.118 |
-0.148 |
| 31 |
245671_at |
AT1G28230
|
ATPUP1, PUP1, purine permease 1 |
0.113 |
0.781 |
| 32 |
265338_at |
AT2G18400
|
[ribosomal protein L6 family protein] |
0.113 |
0.029 |
| 33 |
265663_at |
AT2G24290
|
unknown |
0.112 |
0.618 |
| 34 |
259942_at |
AT1G71260
|
ATWHY2, WHIRLY 2, WHY2, WHIRLY 2 |
0.111 |
1.354 |
| 35 |
264476_at |
AT1G77130
|
GUX3, glucuronic acid substitution of xylan 3, PGSIP2, plant glycogenin-like starch initiation protein 2 |
0.109 |
1.075 |
| 36 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.108 |
-0.719 |
| 37 |
247746_at |
AT5G58970
|
ATUCP2, uncoupling protein 2, UCP2, uncoupling protein 2 |
0.107 |
-0.574 |
| 38 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
0.107 |
-0.565 |
| 39 |
257432_at |
AT2G21850
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.104 |
0.899 |
| 40 |
247616_at |
AT5G60260
|
unknown |
0.104 |
-0.089 |
| 41 |
257193_at |
AT3G13160
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.104 |
0.146 |
| 42 |
251927_at |
AT3G53990
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.103 |
0.607 |
| 43 |
266601_at |
AT2G46060
|
[transmembrane protein-related] |
0.102 |
-0.058 |
| 44 |
253199_at |
AT4G35410
|
[Clathrin adaptor complex small chain family protein] |
0.101 |
-0.313 |
| 45 |
253553_at |
AT4G31050
|
LIP2, octanoyl- transferase |
0.100 |
0.053 |
| 46 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.099 |
0.366 |
| 47 |
259005_at |
AT3G01930
|
[Major facilitator superfamily protein] |
0.098 |
-0.022 |
| 48 |
257317_at |
ATMG01060
|
ORF107G |
0.098 |
-0.957 |
| 49 |
257751_at |
AT3G18690
|
MKS1, MAP kinase substrate 1 |
0.097 |
1.288 |
| 50 |
250412_at |
AT5G11150
|
ATVAMP713, vesicle-associated membrane protein 713, VAMP713, VAMP713, vesicle-associated membrane protein 713 |
0.095 |
0.050 |
| 51 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.095 |
-0.820 |
| 52 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.288 |
0.818 |
| 53 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-0.287 |
0.318 |
| 54 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.256 |
-0.495 |
| 55 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.245 |
-0.575 |
| 56 |
258796_at |
AT3G04630
|
WDL1, WVD2-like 1 |
-0.231 |
0.501 |
| 57 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.222 |
-0.844 |
| 58 |
249837_at |
AT5G23480
|
[SWIB/MDM2 domain] |
-0.217 |
-0.715 |
| 59 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.215 |
-0.782 |
| 60 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
-0.213 |
-0.130 |
| 61 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
-0.200 |
0.135 |
| 62 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.192 |
-0.407 |
| 63 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.190 |
0.275 |
| 64 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.190 |
0.771 |
| 65 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
-0.186 |
-0.692 |
| 66 |
252412_at |
AT3G47295
|
unknown |
-0.185 |
-0.847 |
| 67 |
264331_at |
AT1G04130
|
AtTPR2, TPR2, tetratricopeptide repeat 2 |
-0.184 |
-0.356 |
| 68 |
267141_at |
AT2G38090
|
[Duplicated homeodomain-like superfamily protein] |
-0.173 |
0.882 |
| 69 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.173 |
-0.092 |
| 70 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
-0.169 |
-0.836 |
| 71 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
-0.168 |
0.341 |
| 72 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.165 |
0.570 |
| 73 |
259537_at |
AT1G12370
|
PHR1, photolyase 1, UVR2, UV RESISTANCE 2 |
-0.163 |
0.386 |
| 74 |
245411_at |
AT4G17240
|
unknown |
-0.163 |
0.449 |
| 75 |
245792_at |
AT1G32100
|
ATPRR1, PRR1, pinoresinol reductase 1 |
-0.157 |
0.256 |
| 76 |
264343_at |
AT1G11850
|
unknown |
-0.156 |
0.181 |
| 77 |
246144_at |
AT5G20110
|
[Dynein light chain type 1 family protein] |
-0.156 |
0.180 |
| 78 |
265059_at |
AT1G52080
|
AR791 |
-0.152 |
0.301 |
| 79 |
251049_at |
AT5G02430
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.151 |
0.461 |
| 80 |
250036_at |
AT5G18340
|
[ARM repeat superfamily protein] |
-0.150 |
-0.640 |
| 81 |
245352_at |
AT4G15490
|
UGT84A3 |
-0.150 |
-0.152 |
| 82 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
-0.149 |
-0.189 |
| 83 |
249729_at |
AT5G24410
|
PGL4, 6-phosphogluconolactonase 4 |
-0.146 |
0.612 |
| 84 |
253725_at |
AT4G29340
|
PRF4, profilin 4 |
-0.145 |
-0.494 |
| 85 |
255208_at |
AT4G07580
|
[pseudogene] |
-0.140 |
-0.074 |
| 86 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
-0.139 |
0.720 |
| 87 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
-0.133 |
0.158 |
| 88 |
266015_at |
AT2G24190
|
SDR2, short-chain dehydrogenase/reductase 2 |
-0.132 |
1.126 |
| 89 |
254759_at |
AT4G13180
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.132 |
1.222 |
| 90 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.131 |
-0.527 |
| 91 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.129 |
0.787 |
| 92 |
259083_at |
AT3G04810
|
ATNEK2, NIMA-related kinase 2, NEK2, NIMA-related kinase 2 |
-0.128 |
0.062 |
| 93 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.125 |
-0.106 |
| 94 |
262003_at |
AT1G64460
|
[Protein kinase superfamily protein] |
-0.123 |
-0.890 |
| 95 |
264357_at |
AT1G03360
|
ATRRP4, ribosomal RNA processing 4, RRP4, ribosomal RNA processing 4 |
-0.123 |
0.232 |
| 96 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
-0.122 |
0.991 |
| 97 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
-0.122 |
0.370 |
| 98 |
256415_at |
AT3G11210
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.120 |
-0.814 |
| 99 |
253192_at |
AT4G35370
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.116 |
-1.046 |
| 100 |
266895_at |
AT2G26040
|
PYL2, PYR1-like 2, RCAR14, regulatory components of ABA receptor 14 |
-0.116 |
-0.612 |
| 101 |
251152_at |
AT3G63130
|
ATRANGAP1, RAN GTPASE-ACTIVATING PROTEIN 1, RANGAP1, RAN GTPase activating protein 1 |
-0.116 |
0.140 |