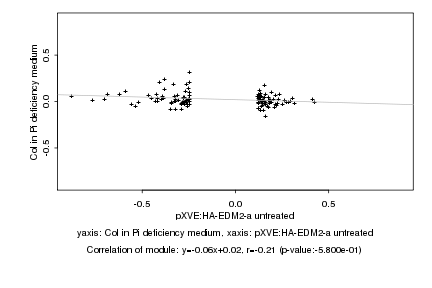
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pXVE:HA-EDM2-a untreated |
Log2 signal ratio
Col in Pi deficiency medium |
| 1 |
245532_at |
AT4G15110
|
CYP97B3, cytochrome P450, family 97, subfamily B, polypeptide 3 |
0.419 |
-0.006 |
| 2 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.409 |
0.030 |
| 3 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.314 |
-0.018 |
| 4 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.301 |
0.041 |
| 5 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.292 |
0.005 |
| 6 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
0.280 |
-0.007 |
| 7 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.269 |
-0.001 |
| 8 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.262 |
0.020 |
| 9 |
266034_at |
AT2G06005
|
FIP1, FRIGIDA interacting protein 1 |
0.252 |
-0.028 |
| 10 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
0.236 |
0.076 |
| 11 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.228 |
0.030 |
| 12 |
261866_at |
AT1G50420
|
SCL-3, SCARECROW-LIKE 3, SCL3, scarecrow-like 3 |
0.225 |
-0.015 |
| 13 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.215 |
-0.041 |
| 14 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.213 |
0.069 |
| 15 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.210 |
-0.023 |
| 16 |
252237_at |
AT3G49950
|
[GRAS family transcription factor] |
0.209 |
-0.060 |
| 17 |
254274_at |
AT4G22770
|
AHL2, AT-hook motif nuclear localized protein 2 |
0.199 |
0.023 |
| 18 |
263183_at |
AT1G05570
|
ATGSL06, ATGSL6, CALS1, callose synthase 1, GSL06, GSL6, GLUCAN SYNTHASE-LIKE 6 |
0.193 |
-0.010 |
| 19 |
254418_at |
AT4G21480
|
STP12, sugar transporter protein 12 |
0.189 |
0.098 |
| 20 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
0.186 |
-0.003 |
| 21 |
245341_at |
AT4G16447
|
unknown |
0.184 |
0.016 |
| 22 |
253269_at |
AT4G34140
|
[D111/G-patch domain-containing protein] |
0.179 |
0.015 |
| 23 |
257852_at |
AT3G12950
|
[Trypsin family protein] |
0.178 |
-0.020 |
| 24 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
0.175 |
0.013 |
| 25 |
250860_at |
AT5G04770
|
ATCAT6, ARABIDOPSIS THALIANA CATIONIC AMINO ACID TRANSPORTER 6, CAT6, cationic amino acid transporter 6 |
0.175 |
-0.064 |
| 26 |
262396_at |
AT1G49470
|
unknown |
0.172 |
0.051 |
| 27 |
261596_at |
AT1G33080
|
[MATE efflux family protein] |
0.171 |
-0.050 |
| 28 |
252427_at |
AT3G47640
|
PYE, POPEYE |
0.159 |
-0.004 |
| 29 |
258005_at |
AT3G19390
|
[Granulin repeat cysteine protease family protein] |
0.158 |
0.079 |
| 30 |
264900_at |
AT1G23080
|
ATPIN7, ARABIDOPSIS PIN-FORMED 7, PIN7, PIN-FORMED 7 |
0.157 |
-0.022 |
| 31 |
247529_at |
AT5G61520
|
[Major facilitator superfamily protein] |
0.156 |
-0.156 |
| 32 |
260462_at |
AT1G10970
|
ATZIP4, ZIP4, zinc transporter 4 precursor |
0.154 |
0.173 |
| 33 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
0.152 |
0.050 |
| 34 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.152 |
0.009 |
| 35 |
246952_at |
AT5G04820
|
ATOFP13, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 13, OFP13, ovate family protein 13 |
0.151 |
-0.020 |
| 36 |
266250_at |
AT2G27870
|
[similar to RNase H domain-containing protein [Arabidopsis thaliana] (TAIR:AT2G22350.1)] |
0.149 |
0.012 |
| 37 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
0.148 |
-0.088 |
| 38 |
248166_at |
AT5G54520
|
[Transducin/WD40 repeat-like superfamily protein] |
0.146 |
0.012 |
| 39 |
252981_at |
AT4G38260
|
unknown |
0.144 |
0.014 |
| 40 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
0.143 |
-0.020 |
| 41 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
0.140 |
-0.040 |
| 42 |
260622_at |
AT1G07980
|
NF-YC10, nuclear factor Y, subunit C10 |
0.139 |
-0.050 |
| 43 |
261048_at |
AT1G01420
|
UGT72B3, UDP-glucosyl transferase 72B3 |
0.137 |
0.001 |
| 44 |
260592_at |
AT1G55850
|
ATCSLE1, CSLE1, cellulose synthase like E1 |
0.136 |
0.056 |
| 45 |
264130_at |
AT1G79160
|
unknown |
0.136 |
0.011 |
| 46 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.132 |
-0.090 |
| 47 |
259723_at |
AT1G60960
|
ATIRT3, IRON REGULATED TRANSPORTER 3, IRT3, iron regulated transporter 3 |
0.131 |
0.094 |
| 48 |
250747_at |
AT5G05900
|
[UDP-Glycosyltransferase superfamily protein] |
0.130 |
0.045 |
| 49 |
252187_at |
AT3G50850
|
[Putative methyltransferase family protein] |
0.130 |
0.012 |
| 50 |
262703_at |
AT1G16510
|
SAUR41, SMALL AUXIN UPREGULATED 41 |
0.127 |
0.123 |
| 51 |
249947_at |
AT5G19200
|
TSC10B, TSC10B |
0.127 |
0.074 |
| 52 |
262407_at |
AT1G34630
|
unknown |
0.124 |
0.026 |
| 53 |
265379_at |
AT2G18340
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
0.124 |
0.030 |
| 54 |
267580_at |
AT2G41990
|
unknown |
0.121 |
0.036 |
| 55 |
246231_at |
AT4G37080
|
unknown |
0.121 |
-0.021 |
| 56 |
253163_at |
AT4G35750
|
[SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein] |
0.120 |
0.081 |
| 57 |
255402_at |
AT4G03205
|
hemf2 |
0.120 |
-0.070 |
| 58 |
250913_at |
AT5G03770
|
AtKdtA, KDTA, KDO transferase A |
0.120 |
0.015 |
| 59 |
248751_at |
AT5G47540
|
[Mo25 family protein] |
0.119 |
-0.002 |
| 60 |
247565_at |
AT5G61150
|
VIP4, VERNALIZATION INDEPENDENCE 4 |
0.119 |
-0.007 |
| 61 |
261968_at |
AT1G65850
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.119 |
-0.068 |
| 62 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.118 |
0.058 |
| 63 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
-0.883 |
0.059 |
| 64 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
-0.767 |
0.019 |
| 65 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.707 |
0.032 |
| 66 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
-0.689 |
0.077 |
| 67 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
-0.622 |
0.085 |
| 68 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
-0.594 |
0.118 |
| 69 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
-0.561 |
-0.022 |
| 70 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
-0.540 |
-0.049 |
| 71 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.520 |
-0.007 |
| 72 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.467 |
0.073 |
| 73 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.453 |
0.042 |
| 74 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.432 |
0.009 |
| 75 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
-0.427 |
0.082 |
| 76 |
252377_at |
AT3G47960
|
AtNPF2.10, GTR1, GLUCOSINOLATE TRANSPORTER-1, NPF2.10, NRT1/ PTR family 2.10 |
-0.421 |
0.002 |
| 77 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.419 |
0.040 |
| 78 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
-0.409 |
0.210 |
| 79 |
248719_at |
AT5G47910
|
ATRBOHD, RBOHD, respiratory burst oxidase homologue D |
-0.399 |
0.026 |
| 80 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.392 |
0.056 |
| 81 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.390 |
0.040 |
| 82 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-0.384 |
0.239 |
| 83 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
-0.381 |
0.140 |
| 84 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.351 |
-0.076 |
| 85 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.346 |
-0.019 |
| 86 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
-0.343 |
-0.008 |
| 87 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
-0.334 |
0.189 |
| 88 |
266299_at |
AT2G29450
|
AT103-1A, ATGSTU1, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 1, ATGSTU5, glutathione S-transferase tau 5, GSTU5, glutathione S-transferase tau 5 |
-0.332 |
0.008 |
| 89 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.327 |
0.060 |
| 90 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
-0.326 |
-0.076 |
| 91 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
-0.322 |
0.007 |
| 92 |
258979_at |
AT3G09440
|
[Heat shock protein 70 (Hsp 70) family protein] |
-0.322 |
0.023 |
| 93 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
-0.315 |
0.069 |
| 94 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
-0.307 |
0.020 |
| 95 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
-0.292 |
-0.084 |
| 96 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.291 |
-0.021 |
| 97 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.290 |
-0.022 |
| 98 |
253654_at |
AT4G30060
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.286 |
-0.004 |
| 99 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.282 |
0.038 |
| 100 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.282 |
-0.019 |
| 101 |
258805_at |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
-0.281 |
-0.010 |
| 102 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
-0.279 |
0.052 |
| 103 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
-0.278 |
-0.027 |
| 104 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.278 |
0.048 |
| 105 |
246888_at |
AT5G26270
|
unknown |
-0.270 |
0.011 |
| 106 |
248821_at |
AT5G47070
|
[Protein kinase superfamily protein] |
-0.270 |
0.109 |
| 107 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
-0.264 |
0.021 |
| 108 |
261506_at |
AT1G71697
|
ATCK1, choline kinase 1, CK, CHOLINE KINASE, CK1, choline kinase 1 |
-0.263 |
0.012 |
| 109 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.263 |
-0.005 |
| 110 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
-0.263 |
0.191 |
| 111 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
-0.259 |
-0.022 |
| 112 |
248123_at |
AT5G54720
|
[Ankyrin repeat family protein] |
-0.259 |
-0.028 |
| 113 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.259 |
-0.043 |
| 114 |
265075_at |
AT1G55450
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.256 |
0.023 |
| 115 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
-0.255 |
0.141 |
| 116 |
251309_at |
AT3G61220
|
SDR1, short-chain dehydrogenase/reductase 1 |
-0.251 |
0.031 |
| 117 |
259479_at |
AT1G19020
|
unknown |
-0.250 |
0.207 |
| 118 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.250 |
0.068 |
| 119 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
-0.250 |
0.103 |
| 120 |
253630_at |
AT4G30490
|
[AFG1-like ATPase family protein] |
-0.250 |
0.041 |
| 121 |
260009_at |
AT1G67950
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.249 |
-0.025 |
| 122 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
-0.249 |
0.316 |
| 123 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
-0.249 |
0.003 |