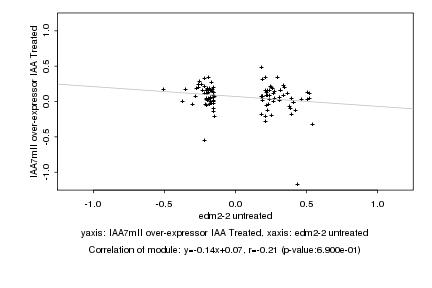
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
edm2-2 untreated |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
0.539 |
-0.324 |
| 2 |
249726_at |
AT5G35480
|
unknown |
0.517 |
0.050 |
| 3 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.516 |
0.127 |
| 4 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
0.506 |
0.037 |
| 5 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.505 |
0.135 |
| 6 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.459 |
0.035 |
| 7 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.433 |
-1.160 |
| 8 |
247640_at |
AT5G60610
|
[F-box/RNI-like superfamily protein] |
0.421 |
-0.127 |
| 9 |
245532_at |
AT4G15110
|
CYP97B3, cytochrome P450, family 97, subfamily B, polypeptide 3 |
0.404 |
-0.001 |
| 10 |
263515_at |
AT2G21640
|
unknown |
0.403 |
-0.011 |
| 11 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.390 |
-0.182 |
| 12 |
248405_at |
AT5G51480
|
SKS2, SKU5 similar 2 |
0.389 |
0.051 |
| 13 |
255148_at |
AT4G08470
|
MAPKKK10, MEKK3, MAPK/ERK kinase kinase 3 |
0.382 |
-0.087 |
| 14 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
0.375 |
-0.070 |
| 15 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.366 |
0.125 |
| 16 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.341 |
0.211 |
| 17 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
0.334 |
0.231 |
| 18 |
264635_at |
AT1G65500
|
unknown |
0.332 |
0.091 |
| 19 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.313 |
0.164 |
| 20 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
0.311 |
0.160 |
| 21 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
0.310 |
0.063 |
| 22 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.309 |
0.026 |
| 23 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.291 |
0.346 |
| 24 |
266139_at |
AT2G28085
|
SAUR42, SMALL AUXIN UPREGULATED RNA 42 |
0.275 |
0.049 |
| 25 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.269 |
0.154 |
| 26 |
261567_at |
AT1G33055
|
unknown |
0.268 |
0.007 |
| 27 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
0.268 |
0.127 |
| 28 |
256062_at |
AT1G07090
|
LSH6, LIGHT SENSITIVE HYPOCOTYLS 6 |
0.259 |
0.192 |
| 29 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.253 |
-0.186 |
| 30 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
0.250 |
0.210 |
| 31 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.240 |
0.220 |
| 32 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
0.237 |
0.170 |
| 33 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.236 |
0.091 |
| 34 |
256188_at |
AT1G30160
|
unknown |
0.233 |
0.041 |
| 35 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.227 |
-0.037 |
| 36 |
248394_at |
AT5G52070
|
[Agenet domain-containing protein] |
0.225 |
-0.126 |
| 37 |
256597_at |
AT3G28500
|
[60S acidic ribosomal protein family] |
0.225 |
0.090 |
| 38 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.220 |
0.165 |
| 39 |
265189_at |
AT1G23840
|
unknown |
0.217 |
0.095 |
| 40 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
0.216 |
0.141 |
| 41 |
258257_at |
AT3G26770
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.214 |
-0.051 |
| 42 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.211 |
0.349 |
| 43 |
266824_at |
AT2G22800
|
HAT9 |
0.210 |
-0.272 |
| 44 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.210 |
0.157 |
| 45 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
0.210 |
-0.207 |
| 46 |
247529_at |
AT5G61520
|
[Major facilitator superfamily protein] |
0.189 |
0.077 |
| 47 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.189 |
0.051 |
| 48 |
256664_at |
AT3G12040
|
[DNA-3-methyladenine glycosylase (MAG)] |
0.188 |
0.019 |
| 49 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.188 |
0.082 |
| 50 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.187 |
0.317 |
| 51 |
261586_at |
AT1G01640
|
[BTB/POZ domain-containing protein] |
0.180 |
-0.183 |
| 52 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.179 |
0.075 |
| 53 |
258203_at |
AT3G13950
|
unknown |
0.178 |
0.486 |
| 54 |
263241_at |
AT2G16500
|
ADC1, arginine decarboxylase 1, ARGDC, ARGDC1, SPE1 |
-0.512 |
0.172 |
| 55 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.374 |
0.014 |
| 56 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
-0.354 |
0.182 |
| 57 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
-0.308 |
-0.042 |
| 58 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
-0.284 |
0.082 |
| 59 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
-0.281 |
0.196 |
| 60 |
252377_at |
AT3G47960
|
AtNPF2.10, GTR1, GLUCOSINOLATE TRANSPORTER-1, NPF2.10, NRT1/ PTR family 2.10 |
-0.264 |
0.244 |
| 61 |
261602_at |
AT1G49630
|
ATPREP2, presequence protease 2, PREP2, presequence protease 2 |
-0.261 |
0.203 |
| 62 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.256 |
0.290 |
| 63 |
248719_at |
AT5G47910
|
ATRBOHD, RBOHD, respiratory burst oxidase homologue D |
-0.245 |
0.242 |
| 64 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.233 |
0.165 |
| 65 |
254408_at |
AT4G21390
|
B120 |
-0.225 |
0.125 |
| 66 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.224 |
-0.540 |
| 67 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
-0.222 |
0.226 |
| 68 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
-0.220 |
0.339 |
| 69 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
-0.217 |
-0.030 |
| 70 |
260206_at |
AT1G70740
|
[Protein kinase superfamily protein] |
-0.209 |
0.122 |
| 71 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
-0.209 |
0.048 |
| 72 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.208 |
0.161 |
| 73 |
253397_at |
AT4G32710
|
PERK14, proline-rich extensin-like receptor kinase 14 |
-0.206 |
-0.050 |
| 74 |
260477_at |
AT1G11050
|
[Protein kinase superfamily protein] |
-0.206 |
0.040 |
| 75 |
263629_at |
AT2G04850
|
[Auxin-responsive family protein] |
-0.203 |
0.120 |
| 76 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
-0.201 |
0.195 |
| 77 |
264227_at |
AT1G67500
|
ATREV3, recovery protein 3, REV3, recovery protein 3 |
-0.198 |
0.017 |
| 78 |
258282_at |
AT3G26910
|
[hydroxyproline-rich glycoprotein family protein] |
-0.197 |
0.179 |
| 79 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.196 |
0.032 |
| 80 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
-0.193 |
0.133 |
| 81 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
-0.192 |
0.052 |
| 82 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.190 |
0.352 |
| 83 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
-0.186 |
-0.035 |
| 84 |
259945_at |
AT1G71460
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.185 |
0.186 |
| 85 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
-0.182 |
0.161 |
| 86 |
263630_at |
AT2G04845
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.179 |
0.048 |
| 87 |
262292_at |
AT1G27595
|
unknown |
-0.178 |
0.064 |
| 88 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
-0.176 |
-0.019 |
| 89 |
258979_at |
AT3G09440
|
[Heat shock protein 70 (Hsp 70) family protein] |
-0.176 |
0.013 |
| 90 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
-0.170 |
0.281 |
| 91 |
267344_at |
AT2G44230
|
unknown |
-0.169 |
-0.024 |
| 92 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.165 |
0.184 |
| 93 |
253643_at |
AT4G29780
|
unknown |
-0.163 |
0.155 |
| 94 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
-0.158 |
-0.091 |
| 95 |
247147_at |
AT5G65630
|
GTE7, global transcription factor group E7 |
-0.158 |
0.060 |
| 96 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.157 |
0.135 |
| 97 |
254181_at |
AT4G23940
|
ARC1, ACCUMULATION AND REPLICATION OF CHLOROPLASTS 1, FtsHi1, FTSH inactive protease 1 |
-0.157 |
-0.024 |
| 98 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
-0.157 |
-0.136 |
| 99 |
260023_at |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
-0.157 |
0.140 |
| 100 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.157 |
0.098 |
| 101 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
-0.156 |
0.212 |
| 102 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.156 |
0.009 |
| 103 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-0.155 |
0.025 |
| 104 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
-0.152 |
-0.198 |
| 105 |
251636_at |
AT3G57530
|
ATCPK32, CDPK32, CPK32, calcium-dependent protein kinase 32 |
-0.152 |
0.084 |