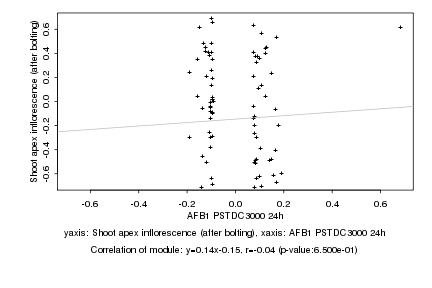
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
AFB1 PSTDC3000 24h |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
255417_at |
AT4G03190
|
AFB1, AUXIN SIGNALING F BOX PROTEIN 1, ATGRH1, GRH1, GRR1-like protein 1 |
0.681 |
0.619 |
| 2 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
0.189 |
-0.596 |
| 3 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
0.175 |
-0.192 |
| 4 |
256210_at |
AT1G50950
|
[INVOLVED IN: cell redox homeostasis] |
0.166 |
0.534 |
| 5 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.166 |
-0.669 |
| 6 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
0.165 |
-0.059 |
| 7 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.165 |
-0.406 |
| 8 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
0.155 |
-0.610 |
| 9 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.147 |
0.237 |
| 10 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
0.147 |
-0.480 |
| 11 |
257139_at |
AT3G28890
|
AtRLP43, receptor like protein 43, RLP43, receptor like protein 43 |
0.140 |
-0.486 |
| 12 |
249092_at |
AT5G43710
|
[Glycosyl hydrolase family 47 protein] |
0.126 |
0.455 |
| 13 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
0.124 |
0.400 |
| 14 |
249763_at |
AT5G24010
|
[Protein kinase superfamily protein] |
0.121 |
0.448 |
| 15 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
0.121 |
0.048 |
| 16 |
262756_at |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
0.117 |
-0.832 |
| 17 |
261485_at |
AT1G14360
|
ATUTR3, UTR3, UDP-galactose transporter 3 |
0.106 |
0.573 |
| 18 |
267164_at |
AT2G37700
|
[Fatty acid hydroxylase superfamily] |
0.105 |
-0.698 |
| 19 |
253687_at |
AT4G29520
|
[LOCATED IN: endoplasmic reticulum, plasma membrane] |
0.104 |
0.136 |
| 20 |
259540_at |
AT1G20640
|
NLP4, NIN-like protein 4 |
0.101 |
-0.389 |
| 21 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
0.100 |
-0.790 |
| 22 |
252800_at |
AT3G42330
|
unknown |
0.097 |
-1.227 |
| 23 |
247155_at |
AT5G65750
|
[2-oxoglutarate dehydrogenase, E1 component] |
0.096 |
-0.618 |
| 24 |
260896_at |
AT1G29310
|
[SecY protein transport family protein] |
0.096 |
0.360 |
| 25 |
250596_at |
AT5G07780
|
AtFH19, FH19, formin homolog 19 |
0.095 |
-1.034 |
| 26 |
261924_at |
AT1G22550
|
[Major facilitator superfamily protein] |
0.092 |
0.114 |
| 27 |
262003_at |
AT1G64460
|
[Protein kinase superfamily protein] |
0.092 |
-0.842 |
| 28 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
0.091 |
0.377 |
| 29 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
0.087 |
-0.295 |
| 30 |
250037_at |
AT5G18350
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.085 |
-0.480 |
| 31 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
0.085 |
0.325 |
| 32 |
250387_at |
AT5G11290
|
unknown |
0.084 |
-0.637 |
| 33 |
258385_at |
AT3G15510
|
ANAC056, Arabidopsis NAC domain containing protein 56, ATNAC2, NAC domain containing protein 2, NAC2, NAC domain containing protein 2, NARS1, NAC-REGULATED SEED MORPHOLOGY 1 |
0.082 |
-0.486 |
| 34 |
267483_at |
AT2G02810
|
ATUTR1, UDP-GALACTOSE TRANSPORTER 1, UTR1, UDP-galactose transporter 1 |
0.081 |
0.845 |
| 35 |
263902_at |
AT2G36230
|
APG10, ALBINO AND PALE GREEN 10, HISN3 |
0.081 |
0.374 |
| 36 |
246632_at |
AT1G29710
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.081 |
-0.511 |
| 37 |
253404_at |
AT4G32840
|
PFK6, phosphofructokinase 6 |
0.079 |
1.204 |
| 38 |
262833_at |
AT1G14750
|
SDS, SOLO DANCERS |
0.078 |
-0.712 |
| 39 |
255460_at |
AT4G02800
|
unknown |
0.078 |
1.255 |
| 40 |
254465_at |
AT4G20420
|
[Tapetum specific protein TAP35/TAP44] |
0.077 |
-0.198 |
| 41 |
266408_at |
AT2G38520
|
[copia-like retrotransposon family, has a 8.4e-60 P-value blast match to dbj|BAA78427.1| polyprotein (AtRE2-2) (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.077 |
-0.506 |
| 42 |
262139_at |
AT1G52610
|
[Mutator-like transposase family, has a 2.0e-18 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.076 |
-0.121 |
| 43 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
0.076 |
-0.263 |
| 44 |
255218_at |
AT4G07700
|
[gypsy-like retrotransposon family (Athila), has a 3.9e-13 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
0.075 |
-0.890 |
| 45 |
263734_at |
AT1G60030
|
ATNAT7, ARABIDOPSIS NUCLEOBASE-ASCORBATE TRANSPORTER 7, NAT7, nucleobase-ascorbate transporter 7 |
0.074 |
-1.096 |
| 46 |
255491_at |
AT4G02670
|
AtIDD12, indeterminate(ID)-domain 12, IDD12, indeterminate(ID)-domain 12 |
0.072 |
0.212 |
| 47 |
255385_at |
AT4G03610
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
0.072 |
-0.133 |
| 48 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.072 |
-0.037 |
| 49 |
258174_at |
AT3G21470
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.072 |
-1.147 |
| 50 |
248231_at |
AT5G53770
|
[Nucleotidyltransferase family protein] |
0.071 |
0.415 |
| 51 |
251946_at |
AT3G53540
|
TRM19, TON1 Recruiting Motif 19 |
0.071 |
0.635 |
| 52 |
250605_at |
AT5G07570
|
[glycine/proline-rich protein] |
0.068 |
-0.783 |
| 53 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.366 |
-0.839 |
| 54 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
-0.193 |
0.245 |
| 55 |
262719_at |
AT1G43590
|
unknown |
-0.190 |
-0.296 |
| 56 |
255726_at |
AT1G25530
|
[Transmembrane amino acid transporter family protein] |
-0.160 |
0.046 |
| 57 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.160 |
0.357 |
| 58 |
249920_at |
AT5G19260
|
FAF3, FANTASTIC FOUR 3 |
-0.149 |
0.621 |
| 59 |
246274_at |
AT4G36620
|
GATA19, GATA transcription factor 19, HANL2, hanaba taranu like 2 |
-0.144 |
-0.707 |
| 60 |
252209_at |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.139 |
-0.053 |
| 61 |
250933_at |
AT5G03170
|
ATFLA11, ARABIDOPSIS FASCICLIN-LIKE ARABINOGALACTAN-PROTEIN 11, FLA11, FASCICLIN-like arabinogalactan-protein 11 |
-0.138 |
-0.454 |
| 62 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.135 |
0.487 |
| 63 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.127 |
0.455 |
| 64 |
253010_at |
AT4G37750
|
ANT, AINTEGUMENTA, CKC, CKC1, COMPLEMENTING A PROTEIN KINASE C MUTANT 1, DRG, DRAGON |
-0.125 |
0.421 |
| 65 |
256602_at |
AT3G28310
|
unknown |
-0.123 |
0.215 |
| 66 |
264520_at |
AT1G10010
|
AAP8, amino acid permease 8, ATAAP8 |
-0.122 |
-0.499 |
| 67 |
262752_at |
AT1G16330
|
CYCB3;1, cyclin b3;1 |
-0.112 |
0.412 |
| 68 |
253419_at |
AT4G32780
|
[phosphoinositide binding] |
-0.111 |
-1.021 |
| 69 |
245875_at |
AT1G26240
|
[Proline-rich extensin-like family protein] |
-0.111 |
0.390 |
| 70 |
261476_at |
AT1G14480
|
[Ankyrin repeat family protein] |
-0.109 |
-0.249 |
| 71 |
260320_at |
AT1G63930
|
ROH1, from the Czech 'roh' meaning 'corner' |
-0.107 |
0.103 |
| 72 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
-0.107 |
-0.802 |
| 73 |
257994_at |
AT3G19920
|
unknown |
-0.107 |
-0.043 |
| 74 |
253008_at |
AT4G38210
|
ATEXP20, ATEXPA20, expansin A20, ATHEXP ALPHA 1.23, EXP20, EXPANSIN 20, EXPA20, expansin A20 |
-0.106 |
-0.075 |
| 75 |
263061_at |
AT2G18190
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.106 |
-0.381 |
| 76 |
245792_at |
AT1G32100
|
ATPRR1, PRR1, pinoresinol reductase 1 |
-0.105 |
-0.007 |
| 77 |
250031_at |
AT5G18240
|
ATMYR1, ARABIDOPSIS MYB-RELATED PROTEIN 1, MYR1, myb-related protein 1 |
-0.105 |
-1.346 |
| 78 |
266346_at |
AT2G01430
|
ATHB-17, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 17, ATHB17, homeobox-leucine zipper protein 17, HB17, homeobox-leucine zipper protein 17 |
-0.105 |
-0.139 |
| 79 |
257764_at |
AT3G23010
|
AtRLP36, receptor like protein 36, RLP36, receptor like protein 36 |
-0.104 |
-0.036 |
| 80 |
248662_at |
AT5G48690
|
unknown |
-0.104 |
-0.298 |
| 81 |
250230_at |
AT5G13900
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.104 |
-1.016 |
| 82 |
261606_at |
AT1G49570
|
[Peroxidase superfamily protein] |
-0.103 |
0.487 |
| 83 |
267221_at |
AT2G02480
|
STI, STICHEL |
-0.103 |
0.138 |
| 84 |
251485_at |
AT3G59550
|
ATRAD21.2, SISTER CHROMATID COHESION 1 PROTEIN 3, ATSYN3, SYN3 |
-0.102 |
0.409 |
| 85 |
247651_at |
AT5G59870
|
HTA6, histone H2A 6 |
-0.102 |
0.696 |
| 86 |
250587_at |
AT5G07640
|
[RING/U-box superfamily protein] |
-0.100 |
-0.637 |
| 87 |
262255_at |
AT1G53790
|
[F-box and associated interaction domains-containing protein] |
-0.100 |
-1.137 |
| 88 |
262620_at |
AT1G06540
|
unknown |
-0.100 |
0.265 |
| 89 |
247397_at |
AT5G62940
|
DOF5.6, DNA BINDING WITH ONE FINGER 5.6, HCA2, HIGH CAMBIAL ACTIVITY2 |
-0.099 |
-0.092 |
| 90 |
259696_at |
AT1G63150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.099 |
0.021 |
| 91 |
251812_at |
AT3G54970
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.098 |
0.025 |
| 92 |
262140_at |
AT1G52470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.097 |
-0.286 |
| 93 |
251511_at |
AT3G59180
|
[Protein with RNI-like/FBD-like domains] |
-0.097 |
-0.893 |
| 94 |
245497_at |
AT4G16460
|
unknown |
-0.097 |
-1.258 |
| 95 |
254070_at |
AT4G25430
|
TRM23, TON1 Recruiting Motif 23 |
-0.096 |
-1.108 |
| 96 |
260506_at |
AT1G47210
|
CYCA3;2, cyclin-dependent protein kinase 3;2 |
-0.096 |
0.660 |
| 97 |
261067_at |
AT1G07460
|
[Concanavalin A-like lectin family protein] |
-0.095 |
-0.086 |
| 98 |
246315_at |
AT3G56870
|
unknown |
-0.095 |
0.355 |
| 99 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
-0.095 |
0.195 |
| 100 |
263977_at |
AT2G42660
|
[Homeodomain-like superfamily protein] |
-0.095 |
0.039 |
| 101 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
-0.095 |
-0.688 |
| 102 |
254515_at |
AT4G20270
|
BAM3, BARELY ANY MERISTEM 3 |
-0.094 |
1.254 |
| 103 |
261550_at |
AT1G63450
|
RHS8, root hair specific 8, XUT1, XYLOGLUCAN-SPECIFIC GALACTURONOSYLTRANSFERASE 1 |
-0.093 |
0.007 |