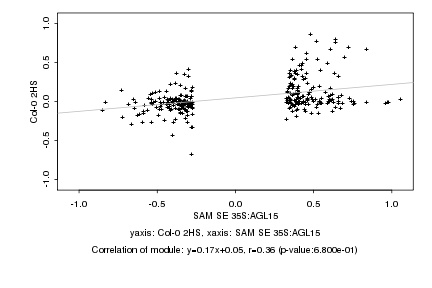
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
SAM SE 35S:AGL15 |
Log2 signal ratio
Col-0 2HS |
| 1 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
1.053 |
0.033 |
| 2 |
253895_at |
AT4G27160
|
AT2S3, SESA3, seed storage albumin 3 |
0.974 |
-0.004 |
| 3 |
249548_at |
AT5G38170
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.971 |
-0.006 |
| 4 |
253894_at |
AT4G27150
|
AT2S2, SESA2, seed storage albumin 2 |
0.956 |
-0.017 |
| 5 |
267317_at |
AT2G34700
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.836 |
-0.001 |
| 6 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.834 |
0.680 |
| 7 |
253902_at |
AT4G27170
|
AT2S4, SESA4, seed storage albumin 4 |
0.760 |
-0.015 |
| 8 |
251202_at |
AT3G63040
|
unknown |
0.754 |
0.001 |
| 9 |
249547_at |
AT5G38160
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.750 |
-0.032 |
| 10 |
267431_at |
AT2G34870
|
MEE26, maternal effect embryo arrest 26 |
0.736 |
0.007 |
| 11 |
253904_at |
AT4G27140
|
AT2S1, SESA1, seed storage albumin 1 |
0.723 |
0.044 |
| 12 |
256442_at |
AT3G10930
|
unknown |
0.719 |
0.696 |
| 13 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
0.695 |
0.568 |
| 14 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.681 |
-0.014 |
| 15 |
250258_at |
AT5G13790
|
AGL15, AGAMOUS-like 15 |
0.672 |
0.080 |
| 16 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
0.670 |
-0.079 |
| 17 |
256290_at |
AT3G12203
|
scpl17, serine carboxypeptidase-like 17 |
0.659 |
0.061 |
| 18 |
261305_at |
AT1G48470
|
GLN1;5, glutamine synthetase 1;5 |
0.655 |
-0.015 |
| 19 |
264314_at |
AT1G70420
|
unknown |
0.655 |
0.327 |
| 20 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.655 |
0.001 |
| 21 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
0.653 |
0.060 |
| 22 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.635 |
0.806 |
| 23 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.634 |
0.762 |
| 24 |
264892_at |
AT1G23160
|
[Auxin-responsive GH3 family protein] |
0.634 |
-0.076 |
| 25 |
265387_at |
AT2G20670
|
unknown |
0.632 |
0.364 |
| 26 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
0.622 |
-0.002 |
| 27 |
253767_at |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
0.619 |
-0.128 |
| 28 |
249939_at |
AT5G22430
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.619 |
0.029 |
| 29 |
267613_at |
AT2G26700
|
PID2, PINOID2 |
0.617 |
0.093 |
| 30 |
248125_at |
AT5G54740
|
SESA5, seed storage albumin 5 |
0.609 |
0.016 |
| 31 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
0.607 |
0.087 |
| 32 |
256645_at |
AT3G24250
|
[glycine-rich protein] |
0.605 |
0.017 |
| 33 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.604 |
0.677 |
| 34 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.602 |
0.169 |
| 35 |
257927_at |
AT3G23240
|
ATERF1, ETHYLENE RESPONSE FACTOR 1, ERF1, ethylene response factor 1 |
0.598 |
0.027 |
| 36 |
249549_at |
AT5G38180
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.597 |
0.004 |
| 37 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
0.593 |
0.070 |
| 38 |
246814_at |
AT5G27200
|
ACP5, acyl carrier protein 5 |
0.593 |
0.002 |
| 39 |
252834_at |
AT4G40070
|
[RING/U-box superfamily protein] |
0.588 |
0.500 |
| 40 |
264187_at |
AT1G54860
|
[Glycoprotein membrane precursor GPI-anchored] |
0.587 |
-0.013 |
| 41 |
258327_at |
AT3G22640
|
PAP85 |
0.572 |
0.126 |
| 42 |
253930_at |
AT4G26740
|
AtCLO1, ATPXG1, ARABIDOPSIS THALIANA PEROXYGENASE 1, ATS1, seed gene 1, CLO1, CALEOSIN1 |
0.547 |
0.039 |
| 43 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
0.546 |
-0.020 |
| 44 |
250501_at |
AT5G09640
|
SCPL19, serine carboxypeptidase-like 19, SNG2, SINAPOYLGLUCOSE ACCUMULATOR 2 |
0.541 |
-0.019 |
| 45 |
248435_at |
AT5G51210
|
OLEO3, oleosin3 |
0.540 |
-0.009 |
| 46 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.538 |
0.403 |
| 47 |
264606_at |
AT1G04660
|
[glycine-rich protein] |
0.533 |
0.002 |
| 48 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.533 |
0.197 |
| 49 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.532 |
0.012 |
| 50 |
248028_at |
AT5G55620
|
unknown |
0.530 |
-0.142 |
| 51 |
253859_at |
AT4G27657
|
unknown |
0.523 |
0.541 |
| 52 |
263175_at |
AT1G05510
|
OBAP1a, Oil Body-Associated Protein 1a |
0.523 |
-0.028 |
| 53 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.513 |
0.773 |
| 54 |
264282_at |
AT1G61840
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.512 |
-0.070 |
| 55 |
265891_at |
AT2G15010
|
[Plant thionin] |
0.511 |
0.033 |
| 56 |
258240_at |
AT3G27660
|
OLE3, OLEOSIN 3, OLEO4, oleosin 4 |
0.499 |
0.001 |
| 57 |
253940_at |
AT4G26950
|
unknown |
0.497 |
0.186 |
| 58 |
265095_at |
AT1G03880
|
CRB, CRUCIFERIN B, CRU2, cruciferin 2 |
0.488 |
0.038 |
| 59 |
251423_at |
AT3G60550
|
CYCP3;2, cyclin p3;2 |
0.486 |
0.165 |
| 60 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
0.486 |
-0.149 |
| 61 |
248754_at |
AT5G47670
|
L1L, LEC1-LIKE, NF-YB6, nuclear factor Y, subunit B6 |
0.484 |
0.052 |
| 62 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.478 |
0.867 |
| 63 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
0.473 |
0.123 |
| 64 |
265468_at |
AT2G37210
|
LOG3, LONELY GUY 3 |
0.469 |
0.116 |
| 65 |
250519_at |
AT5G08460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.467 |
-0.037 |
| 66 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.463 |
0.093 |
| 67 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.459 |
0.237 |
| 68 |
260376_at |
AT1G74110
|
CYP78A10, cytochrome P450, family 78, subfamily A, polypeptide 10 |
0.458 |
0.018 |
| 69 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.455 |
0.029 |
| 70 |
263138_at |
AT1G65090
|
unknown |
0.455 |
0.041 |
| 71 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.454 |
0.618 |
| 72 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
0.453 |
0.372 |
| 73 |
247047_at |
AT5G66650
|
unknown |
0.451 |
0.540 |
| 74 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
0.448 |
-0.027 |
| 75 |
245172_at |
AT2G47540
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.446 |
-0.128 |
| 76 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.444 |
0.304 |
| 77 |
249913_at |
AT5G22810
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.444 |
-0.004 |
| 78 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
0.438 |
-0.095 |
| 79 |
247421_at |
AT5G62800
|
[Protein with RING/U-box and TRAF-like domains] |
0.434 |
-0.005 |
| 80 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.433 |
0.073 |
| 81 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.430 |
0.289 |
| 82 |
263377_at |
AT2G40220
|
ABI4, ABA INSENSITIVE 4, ATABI4, GIN6, GLUCOSE INSENSITIVE 6, ISI3, IMPAIRED SUCROSE INDUCTION 3, SAN5, SALOBRENO 5, SIS5, SUGAR-INSENSITIVE 5, SUN6, SUCROSE UNCOUPLED 6 |
0.429 |
0.012 |
| 83 |
259015_at |
AT3G07350
|
unknown |
0.428 |
0.494 |
| 84 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.425 |
0.322 |
| 85 |
247052_at |
AT5G66700
|
ATHB53, ARABIDOPSIS THALIANA HOMEOBOX 53, HB-8, HOMEOBOX-8, HB53, homeobox 53 |
0.423 |
0.073 |
| 86 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
0.420 |
-0.016 |
| 87 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.417 |
0.468 |
| 88 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.417 |
0.416 |
| 89 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.413 |
0.039 |
| 90 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.407 |
0.118 |
| 91 |
265276_at |
AT2G28400
|
unknown |
0.407 |
0.467 |
| 92 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.405 |
0.005 |
| 93 |
264988_at |
AT1G27140
|
ATGSTU14, glutathione S-transferase tau 14, GST13, GLUTATHIONE S-TRANSFERASE 13, GSTU14, glutathione S-transferase tau 14 |
0.402 |
-0.021 |
| 94 |
250620_at |
AT5G07190
|
ATS3, seed gene 3 |
0.400 |
0.057 |
| 95 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.399 |
0.205 |
| 96 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
0.399 |
0.006 |
| 97 |
246063_at |
AT5G19340
|
unknown |
0.398 |
0.127 |
| 98 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
0.397 |
-0.031 |
| 99 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.393 |
-0.092 |
| 100 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.393 |
0.098 |
| 101 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
0.393 |
0.349 |
| 102 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.392 |
0.053 |
| 103 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
0.392 |
0.154 |
| 104 |
251491_at |
AT3G59480
|
[pfkB-like carbohydrate kinase family protein] |
0.391 |
0.174 |
| 105 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
0.390 |
-0.105 |
| 106 |
262828_at |
AT1G14950
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.390 |
-0.005 |
| 107 |
250780_at |
AT5G05290
|
ATEXP2, ATEXPA2, expansin A2, ATHEXP ALPHA 1.12, EXP2, EXPANSIN 2, EXPA2, expansin A2 |
0.389 |
0.006 |
| 108 |
265184_at |
AT1G23710
|
unknown |
0.388 |
0.399 |
| 109 |
247424_at |
AT5G62850
|
AtSWEET5, AtVEX1, VEGETATIVE CELL EXPRESSED1, SWEET5 |
0.386 |
0.029 |
| 110 |
258178_at |
AT3G21680
|
unknown |
0.385 |
-0.180 |
| 111 |
259979_at |
AT1G76600
|
unknown |
0.383 |
0.697 |
| 112 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.381 |
0.362 |
| 113 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
0.379 |
-0.034 |
| 114 |
246891_at |
AT5G25490
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
0.379 |
0.112 |
| 115 |
257321_at |
ATMG01130
|
ORF106F |
0.375 |
0.297 |
| 116 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
0.374 |
0.017 |
| 117 |
249756_at |
AT5G24313
|
unknown |
0.374 |
-0.053 |
| 118 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.373 |
0.395 |
| 119 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.372 |
0.290 |
| 120 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
0.366 |
0.205 |
| 121 |
260435_at |
AT1G68320
|
AtMYB62, myb domain protein 62, BW62B, BW62C, MYB62, myb domain protein 62 |
0.365 |
0.092 |
| 122 |
256452_at |
AT1G75240
|
AtHB33, homeobox protein 33, HB33, homeobox protein 33, ZHD5, zinc-finger homeodomain 5 |
0.363 |
-0.121 |
| 123 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.363 |
0.542 |
| 124 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
0.363 |
0.169 |
| 125 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
0.362 |
0.221 |
| 126 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
0.362 |
0.085 |
| 127 |
261503_at |
AT1G71691
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.361 |
-0.041 |
| 128 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
0.361 |
-0.034 |
| 129 |
250732_at |
AT5G06480
|
[Immunoglobulin E-set superfamily protein] |
0.358 |
-0.018 |
| 130 |
252483_at |
AT3G46600
|
[GRAS family transcription factor] |
0.357 |
0.337 |
| 131 |
246889_at |
AT5G25470
|
[AP2/B3-like transcriptional factor family protein] |
0.352 |
0.028 |
| 132 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
0.351 |
0.406 |
| 133 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
0.348 |
0.239 |
| 134 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
0.347 |
-0.075 |
| 135 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
0.347 |
0.018 |
| 136 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
0.346 |
0.105 |
| 137 |
263937_at |
AT2G35910
|
[RING/U-box superfamily protein] |
0.346 |
0.334 |
| 138 |
266259_at |
AT2G27830
|
unknown |
0.346 |
0.217 |
| 139 |
259964_at |
AT1G53680
|
ATGSTU28, glutathione S-transferase TAU 28, GSTU28, glutathione S-transferase TAU 28 |
0.345 |
-0.087 |
| 140 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
0.345 |
0.361 |
| 141 |
258436_at |
AT3G16720
|
ATL2, TOXICOS EN LEVADURA 2, TL2, TOXICOS EN LEVADURA 2 |
0.344 |
0.317 |
| 142 |
251792_at |
AT3G55550
|
LecRK-S.4, L-type lectin receptor kinase S.4 |
0.340 |
0.042 |
| 143 |
245662_at |
AT1G28190
|
unknown |
0.340 |
0.202 |
| 144 |
266423_at |
AT2G41340
|
RPB5D, RNA polymerase II fifth largest subunit, D |
0.340 |
-0.012 |
| 145 |
248539_at |
AT5G50130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.338 |
-0.017 |
| 146 |
248618_at |
AT5G49620
|
AtMYB78, myb domain protein 78, MYB78, myb domain protein 78 |
0.335 |
0.056 |
| 147 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
0.333 |
0.173 |
| 148 |
259682_at |
AT1G63040
|
[pseudogene] |
0.332 |
0.021 |
| 149 |
258225_at |
AT3G15630
|
unknown |
0.331 |
0.135 |
| 150 |
256455_at |
AT1G75190
|
unknown |
0.330 |
0.121 |
| 151 |
259782_at |
AT1G29680
|
unknown |
0.324 |
0.047 |
| 152 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.322 |
-0.219 |
| 153 |
246626_at |
AT1G48870
|
[Transducin/WD40 repeat-like superfamily protein] |
0.321 |
0.000 |
| 154 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
-0.856 |
-0.114 |
| 155 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
-0.836 |
-0.009 |
| 156 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.732 |
0.149 |
| 157 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.723 |
-0.205 |
| 158 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-0.690 |
-0.035 |
| 159 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.668 |
-0.295 |
| 160 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
-0.654 |
0.038 |
| 161 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.650 |
-0.079 |
| 162 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
-0.645 |
-0.011 |
| 163 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
-0.628 |
-0.177 |
| 164 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.617 |
-0.156 |
| 165 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
-0.595 |
-0.260 |
| 166 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-0.591 |
-0.117 |
| 167 |
262396_at |
AT1G49470
|
unknown |
-0.588 |
-0.145 |
| 168 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
-0.582 |
-0.045 |
| 169 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.566 |
-0.105 |
| 170 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.562 |
0.046 |
| 171 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.545 |
0.091 |
| 172 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.540 |
0.032 |
| 173 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.539 |
-0.018 |
| 174 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.538 |
-0.259 |
| 175 |
256569_at |
AT3G19550
|
unknown |
-0.538 |
-0.006 |
| 176 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.536 |
-0.026 |
| 177 |
250438_at |
AT5G10580
|
unknown |
-0.531 |
0.114 |
| 178 |
262739_at |
AT1G28650
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.525 |
-0.009 |
| 179 |
255008_at |
AT4G10060
|
[Beta-glucosidase, GBA2 type family protein] |
-0.516 |
0.005 |
| 180 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
-0.514 |
0.128 |
| 181 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
-0.510 |
-0.069 |
| 182 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-0.497 |
-0.169 |
| 183 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.492 |
0.131 |
| 184 |
246595_at |
AT5G14780
|
FDH, formate dehydrogenase |
-0.489 |
-0.090 |
| 185 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-0.486 |
0.044 |
| 186 |
249271_at |
AT5G41790
|
CIP1, COP1-interactive protein 1 |
-0.483 |
-0.010 |
| 187 |
249378_at |
AT5G40450
|
unknown |
-0.483 |
-0.059 |
| 188 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
-0.478 |
-0.091 |
| 189 |
252327_at |
AT3G48740
|
AtSWEET11, SWEET11 |
-0.475 |
-0.033 |
| 190 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
-0.463 |
-0.002 |
| 191 |
245075_at |
AT2G23180
|
CYP96A1, cytochrome P450, family 96, subfamily A, polypeptide 1 |
-0.460 |
-0.241 |
| 192 |
247091_at |
AT5G66390
|
PRX72, PEROXIDASE 72 |
-0.456 |
0.060 |
| 193 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.447 |
0.132 |
| 194 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.447 |
-0.033 |
| 195 |
249791_at |
AT5G23810
|
AAP7, amino acid permease 7 |
-0.439 |
0.034 |
| 196 |
251646_at |
AT3G57780
|
unknown |
-0.439 |
-0.064 |
| 197 |
263227_at |
AT1G30750
|
unknown |
-0.435 |
-0.003 |
| 198 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.434 |
0.025 |
| 199 |
267127_at |
AT2G23610
|
ATMES3, ARABIDOPSIS THALIANA METHYL ESTERASE 3, MES3, methyl esterase 3 |
-0.432 |
-0.005 |
| 200 |
260941_at |
AT1G44970
|
[Peroxidase superfamily protein] |
-0.427 |
-0.078 |
| 201 |
263121_at |
AT1G78530
|
[Protein kinase superfamily protein] |
-0.425 |
-0.054 |
| 202 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
-0.425 |
-0.034 |
| 203 |
266209_at |
AT2G27550
|
ATC, centroradialis |
-0.421 |
0.027 |
| 204 |
258017_at |
AT3G19370
|
unknown |
-0.418 |
-0.104 |
| 205 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
-0.418 |
0.231 |
| 206 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.416 |
0.045 |
| 207 |
253525_at |
AT4G31330
|
unknown |
-0.415 |
-0.101 |
| 208 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
-0.408 |
-0.429 |
| 209 |
266325_at |
AT2G46630
|
unknown |
-0.408 |
-0.004 |
| 210 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.404 |
0.006 |
| 211 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
-0.404 |
0.027 |
| 212 |
253052_at |
AT4G37310
|
CYP81H1, cytochrome P450, family 81, subfamily H, polypeptide 1 |
-0.402 |
-0.021 |
| 213 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
-0.401 |
-0.048 |
| 214 |
261070_at |
AT1G07390
|
AtRLP1, receptor like protein 1, RLP1, receptor like protein 1 |
-0.399 |
-0.262 |
| 215 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
-0.396 |
0.014 |
| 216 |
265176_at |
AT1G23520
|
unknown |
-0.388 |
0.045 |
| 217 |
249136_at |
AT5G43180
|
unknown |
-0.387 |
-0.224 |
| 218 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.386 |
0.237 |
| 219 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-0.386 |
-0.088 |
| 220 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
-0.380 |
0.013 |
| 221 |
249231_at |
AT5G42030
|
ABIL4, ABL interactor-like protein 4 |
-0.379 |
0.039 |
| 222 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.378 |
0.364 |
| 223 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
-0.371 |
-0.038 |
| 224 |
260429_at |
AT1G72450
|
JAZ6, jasmonate-zim-domain protein 6, TIFY11B, TIFY DOMAIN PROTEIN 11B |
-0.370 |
0.002 |
| 225 |
262154_at |
AT1G52700
|
[alpha/beta-Hydrolases superfamily protein] |
-0.369 |
0.006 |
| 226 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.368 |
-0.098 |
| 227 |
253246_at |
AT4G34600
|
unknown |
-0.367 |
0.002 |
| 228 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
-0.366 |
0.036 |
| 229 |
253985_at |
AT4G26220
|
CCoAOMT7, caffeoyl coenzyme A ester O-methyltransferase 7 |
-0.366 |
-0.034 |
| 230 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.364 |
-0.101 |
| 231 |
258769_at |
AT3G10870
|
ATMES17, ARABIDOPSIS THALIANA METHYL ESTERASE 17, MES17, methyl esterase 17 |
-0.363 |
0.030 |
| 232 |
253800_at |
AT4G28160
|
[hydroxyproline-rich glycoprotein family protein] |
-0.360 |
-0.008 |
| 233 |
252462_at |
AT3G47250
|
unknown |
-0.356 |
-0.048 |
| 234 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.354 |
0.081 |
| 235 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
-0.352 |
-0.153 |
| 236 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.352 |
-0.139 |
| 237 |
264505_at |
AT1G09380
|
UMAMIT25, Usually multiple acids move in and out Transporters 25 |
-0.352 |
0.016 |
| 238 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.352 |
0.215 |
| 239 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.351 |
-0.039 |
| 240 |
261409_at |
AT1G07640
|
OBP2, URP3, UAS-TAGGED ROOT PATTERNING3 |
-0.350 |
-0.061 |
| 241 |
266356_at |
AT2G32300
|
UCC1, uclacyanin 1 |
-0.350 |
0.083 |
| 242 |
265186_at |
AT1G23560
|
unknown |
-0.346 |
-0.026 |
| 243 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
-0.346 |
-0.081 |
| 244 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.342 |
-0.063 |
| 245 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.341 |
-0.006 |
| 246 |
247357_at |
AT5G63710
|
[Leucine-rich repeat protein kinase family protein] |
-0.339 |
-0.148 |
| 247 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
-0.338 |
-0.048 |
| 248 |
256870_at |
AT3G26300
|
CYP71B34, cytochrome P450, family 71, subfamily B, polypeptide 34 |
-0.337 |
-0.047 |
| 249 |
262680_at |
AT1G75880
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.337 |
-0.079 |
| 250 |
251672_at |
AT3G57230
|
AGL16, AGAMOUS-like 16 |
-0.336 |
-0.045 |
| 251 |
266849_at |
AT2G25940
|
ALPHA-VPE, alpha-vacuolar processing enzyme, ALPHAVPE |
-0.336 |
0.007 |
| 252 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
-0.335 |
-0.012 |
| 253 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.334 |
0.007 |
| 254 |
245352_at |
AT4G15490
|
UGT84A3 |
-0.333 |
-0.029 |
| 255 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.332 |
0.359 |
| 256 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
-0.329 |
0.019 |
| 257 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.328 |
-0.018 |
| 258 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.327 |
-0.000 |
| 259 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
-0.327 |
-0.092 |
| 260 |
263284_at |
AT2G36100
|
CASP1, Casparian strip membrane domain protein 1 |
-0.326 |
-0.030 |
| 261 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-0.323 |
0.073 |
| 262 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
-0.322 |
0.173 |
| 263 |
251188_at |
AT3G62730
|
unknown |
-0.321 |
0.002 |
| 264 |
253779_at |
AT4G28490
|
HAE, HAESA, RLK5, RECEPTOR-LIKE PROTEIN KINASE 5 |
-0.320 |
-0.218 |
| 265 |
255032_at |
AT4G09500
|
[UDP-Glycosyltransferase superfamily protein] |
-0.320 |
-0.091 |
| 266 |
245891_at |
AT5G09220
|
AAP2, amino acid permease 2 |
-0.320 |
0.005 |
| 267 |
257181_at |
AT3G13190
|
unknown |
-0.317 |
-0.127 |
| 268 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.313 |
0.065 |
| 269 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.312 |
-0.008 |
| 270 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.312 |
-0.002 |
| 271 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
-0.311 |
-0.029 |
| 272 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.311 |
-0.117 |
| 273 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.311 |
-0.056 |
| 274 |
262921_at |
AT1G79430
|
APL, ALTERED PHLOEM DEVELOPMENT, WDY, WOODY |
-0.309 |
0.068 |
| 275 |
246566_at |
AT5G14940
|
[Major facilitator superfamily protein] |
-0.309 |
-0.174 |
| 276 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.309 |
-0.006 |
| 277 |
261845_at |
AT1G15960
|
ATNRAMP6, NRAMP6, NRAMP metal ion transporter 6 |
-0.308 |
0.011 |
| 278 |
266625_at |
AT2G35380
|
[Peroxidase superfamily protein] |
-0.308 |
-0.269 |
| 279 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
-0.305 |
-0.024 |
| 280 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
-0.305 |
-0.062 |
| 281 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.303 |
-0.065 |
| 282 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
-0.303 |
0.413 |
| 283 |
258807_at |
AT3G04030
|
MYR2 |
-0.302 |
0.323 |
| 284 |
266172_at |
AT2G39010
|
PIP2;6, PLASMA MEMBRANE INTRINSIC PROTEIN 2;6, PIP2E, plasma membrane intrinsic protein 2E |
-0.301 |
-0.021 |
| 285 |
267459_at |
AT2G33850
|
unknown |
-0.297 |
-0.016 |
| 286 |
249126_at |
AT5G43380
|
TOPP6, type one serine/threonine protein phosphatase 6 |
-0.295 |
-0.018 |
| 287 |
259544_at |
AT1G20620
|
ATCAT3, CAT3, catalase 3, SEN2, SENESCENCE 2 |
-0.293 |
-0.041 |
| 288 |
263668_at |
AT1G04350
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.291 |
0.048 |
| 289 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
-0.291 |
-0.006 |
| 290 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
-0.287 |
0.074 |
| 291 |
248263_at |
AT5G53370
|
ATPMEPCRF, PECTIN METHYLESTERASE PCR FRAGMENT F, PMEPCRF, pectin methylesterase PCR fragment F |
-0.285 |
-0.037 |
| 292 |
262549_at |
AT1G31290
|
AGO3, ARGONAUTE 3 |
-0.284 |
0.144 |
| 293 |
260237_at |
AT1G74430
|
ATMYB95, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 95, ATMYBCP66, ARABIDOPSIS THALIANA MYB DOMAIN CONTAINING PROTEIN 66, MYB95, myb domain protein 95 |
-0.284 |
-0.067 |
| 294 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.283 |
-0.330 |
| 295 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
-0.283 |
-0.024 |
| 296 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
-0.282 |
-0.674 |
| 297 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
-0.282 |
0.132 |
| 298 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
-0.282 |
0.067 |
| 299 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.280 |
-0.332 |
| 300 |
258293_at |
AT3G23430
|
ATPHO1, ARABIDOPSIS PHOSPHATE 1, PHO1, phosphate 1 |
-0.280 |
-0.070 |
| 301 |
246230_at |
AT4G36710
|
AtHAM4, Arabidopsis thaliana HAIRY MERISTEM 4, HAM4 |
-0.279 |
-0.078 |
| 302 |
259116_at |
AT3G01350
|
[Major facilitator superfamily protein] |
-0.279 |
0.184 |
| 303 |
250499_at |
AT5G09730
|
ATBX3, ATBXL3, BETA-XYLOSIDASE 3, BX3, BXL3, beta-xylosidase 3, XYL3 |
-0.278 |
-0.003 |
| 304 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
-0.278 |
-0.037 |
| 305 |
246231_at |
AT4G37080
|
unknown |
-0.277 |
-0.160 |