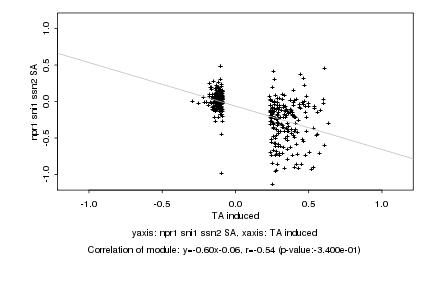
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
TA induced |
Log2 signal ratio
npr1 sni1 ssn2 SA |
| 1 |
249522_at |
AT5G38700
|
unknown |
0.633 |
-0.295 |
| 2 |
261247_at |
AT1G20070
|
unknown |
0.606 |
0.455 |
| 3 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.604 |
-0.599 |
| 4 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.601 |
-0.020 |
| 5 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.599 |
0.031 |
| 6 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.574 |
-0.118 |
| 7 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.567 |
-0.700 |
| 8 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
0.560 |
-0.142 |
| 9 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.557 |
-0.451 |
| 10 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.552 |
-0.462 |
| 11 |
255532_at |
AT4G02170
|
unknown |
0.537 |
-0.057 |
| 12 |
246270_at |
AT4G36500
|
unknown |
0.537 |
-0.084 |
| 13 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.532 |
-0.369 |
| 14 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.527 |
-0.891 |
| 15 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.517 |
-0.921 |
| 16 |
261984_at |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
0.505 |
-0.696 |
| 17 |
258388_at |
AT3G15370
|
ATEXP12, ATEXPA12, expansin 12, ATHEXP ALPHA 1.24, EXP12, EXPANSIN 12, EXPA12, expansin 12 |
0.498 |
-0.066 |
| 18 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.494 |
-0.017 |
| 19 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.485 |
-0.209 |
| 20 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.482 |
0.072 |
| 21 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.482 |
-0.399 |
| 22 |
265075_at |
AT1G55450
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.477 |
-0.139 |
| 23 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.475 |
-0.727 |
| 24 |
245738_at |
AT1G44130
|
[Eukaryotic aspartyl protease family protein] |
0.473 |
-0.057 |
| 25 |
252988_at |
AT4G38410
|
[Dehydrin family protein] |
0.470 |
0.222 |
| 26 |
253937_at |
AT4G26890
|
MAPKKK16, mitogen-activated protein kinase kinase kinase 16 |
0.467 |
0.005 |
| 27 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.461 |
0.321 |
| 28 |
265732_at |
AT2G01300
|
unknown |
0.460 |
-0.537 |
| 29 |
266196_at |
AT2G39110
|
[Protein kinase superfamily protein] |
0.459 |
-0.030 |
| 30 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.457 |
-0.507 |
| 31 |
256186_at |
AT1G51680
|
4CL.1, 4-COUMARATE:COA LIGASE 1, 4CL1, 4-coumarate:CoA ligase 1, AT4CL1, ARABIDOPSIS THALIANA 4-COUMARATE:COA LIGASE 1 |
0.452 |
-0.004 |
| 32 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.450 |
-0.858 |
| 33 |
253799_at |
AT4G28140
|
[Integrase-type DNA-binding superfamily protein] |
0.442 |
0.381 |
| 34 |
253050_at |
AT4G37450
|
AGP18, arabinogalactan protein 18, ATAGP18 |
0.438 |
-0.086 |
| 35 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.435 |
-0.080 |
| 36 |
250464_at |
AT5G10040
|
unknown |
0.434 |
-0.034 |
| 37 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.428 |
-0.908 |
| 38 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.427 |
-0.254 |
| 39 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.419 |
-0.725 |
| 40 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
0.417 |
-0.418 |
| 41 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.415 |
-0.587 |
| 42 |
255756_at |
AT1G19940
|
AtGH9B5, glycosyl hydrolase 9B5, GH9B5, glycosyl hydrolase 9B5 |
0.411 |
0.036 |
| 43 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.411 |
-0.855 |
| 44 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.409 |
-0.300 |
| 45 |
252882_at |
AT4G39675
|
unknown |
0.408 |
-0.218 |
| 46 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
0.407 |
-0.377 |
| 47 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.400 |
0.018 |
| 48 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.400 |
-0.401 |
| 49 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
0.400 |
-0.193 |
| 50 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.399 |
-0.903 |
| 51 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.398 |
-0.213 |
| 52 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.397 |
-0.492 |
| 53 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
0.395 |
-0.007 |
| 54 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.392 |
-0.459 |
| 55 |
245226_at |
AT3G29970
|
[B12D protein] |
0.391 |
-0.054 |
| 56 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
0.391 |
0.156 |
| 57 |
252624_at |
AT3G44735
|
ATPSK3, PHYTOSULFOKINE 3 PRECURSOR, PSK1, PSK3, PHYTOSULFOKINE 3 PRECURSOR |
0.387 |
-0.168 |
| 58 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.387 |
-0.139 |
| 59 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.386 |
-0.187 |
| 60 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.386 |
-0.739 |
| 61 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
0.380 |
-0.116 |
| 62 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.378 |
-0.425 |
| 63 |
256525_at |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
0.377 |
-0.380 |
| 64 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.373 |
-0.627 |
| 65 |
253534_at |
AT4G31500
|
ATR4, ALTERED TRYPTOPHAN REGULATION 4, CYP83B1, cytochrome P450, family 83, subfamily B, polypeptide 1, RED1, RED ELONGATED 1, RNT1, RUNT 1, SUR2, SUPERROOT 2 |
0.373 |
0.020 |
| 66 |
257925_at |
AT3G23170
|
unknown |
0.372 |
-0.104 |
| 67 |
265648_at |
AT2G27500
|
[Glycosyl hydrolase superfamily protein] |
0.372 |
-0.149 |
| 68 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
0.371 |
0.015 |
| 69 |
253632_at |
AT4G30430
|
TET9, tetraspanin9 |
0.359 |
-0.154 |
| 70 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
0.358 |
-0.333 |
| 71 |
253830_at |
AT4G27652
|
unknown |
0.357 |
-0.405 |
| 72 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.355 |
-0.301 |
| 73 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.354 |
-0.534 |
| 74 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
0.354 |
-0.130 |
| 75 |
260367_at |
AT1G69760
|
unknown |
0.352 |
-0.374 |
| 76 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.350 |
-0.404 |
| 77 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.350 |
-0.787 |
| 78 |
263182_at |
AT1G05575
|
unknown |
0.349 |
-0.655 |
| 79 |
264580_at |
AT1G05340
|
unknown |
0.349 |
-0.479 |
| 80 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.348 |
-0.184 |
| 81 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.347 |
-0.120 |
| 82 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.346 |
-0.359 |
| 83 |
247024_at |
AT5G66985
|
unknown |
0.342 |
-0.100 |
| 84 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
0.341 |
-0.215 |
| 85 |
252652_at |
AT3G44720
|
ADT4, arogenate dehydratase 4 |
0.341 |
-0.058 |
| 86 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.340 |
-0.500 |
| 87 |
249910_at |
AT5G22630
|
ADT5, arogenate dehydratase 5 |
0.337 |
-0.132 |
| 88 |
264624_at |
AT1G08930
|
ERD6, EARLY RESPONSE TO DEHYDRATION 6 |
0.335 |
-0.116 |
| 89 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
0.334 |
-0.169 |
| 90 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.334 |
-0.405 |
| 91 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
0.333 |
0.096 |
| 92 |
253643_at |
AT4G29780
|
unknown |
0.332 |
-0.909 |
| 93 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.328 |
-0.073 |
| 94 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.328 |
-0.214 |
| 95 |
259566_at |
AT1G20520
|
unknown |
0.328 |
-0.046 |
| 96 |
250702_at |
AT5G06730
|
[Peroxidase superfamily protein] |
0.324 |
-0.072 |
| 97 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.324 |
-0.351 |
| 98 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.322 |
-0.618 |
| 99 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.321 |
-0.322 |
| 100 |
245363_at |
AT4G15120
|
[VQ motif-containing protein] |
0.317 |
0.099 |
| 101 |
266124_at |
AT2G45080
|
cycp3;1, cyclin p3;1 |
0.316 |
0.034 |
| 102 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.315 |
-0.712 |
| 103 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
0.315 |
-0.346 |
| 104 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.310 |
-0.112 |
| 105 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
0.308 |
-0.312 |
| 106 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.306 |
-0.410 |
| 107 |
261193_at |
AT1G32920
|
unknown |
0.303 |
-0.215 |
| 108 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.301 |
-0.604 |
| 109 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.300 |
-0.054 |
| 110 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.299 |
-0.728 |
| 111 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.299 |
-0.707 |
| 112 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.296 |
-0.078 |
| 113 |
250910_at |
AT5G03720
|
AT-HSFA3, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A3, HSFA3, heat shock transcription factor A3 |
0.294 |
0.047 |
| 114 |
258251_at |
AT3G15810
|
unknown |
0.293 |
-0.097 |
| 115 |
261506_at |
AT1G71697
|
ATCK1, choline kinase 1, CK, CHOLINE KINASE, CK1, choline kinase 1 |
0.292 |
-0.137 |
| 116 |
267293_at |
AT2G23810
|
TET8, tetraspanin8 |
0.292 |
-0.292 |
| 117 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.291 |
-0.613 |
| 118 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.290 |
-0.230 |
| 119 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.290 |
-0.404 |
| 120 |
261567_at |
AT1G33055
|
unknown |
0.290 |
-0.437 |
| 121 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
0.289 |
-0.297 |
| 122 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.288 |
-0.576 |
| 123 |
252557_at |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
0.286 |
-0.275 |
| 124 |
248821_at |
AT5G47070
|
[Protein kinase superfamily protein] |
0.286 |
-0.170 |
| 125 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.286 |
-0.005 |
| 126 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.285 |
-0.319 |
| 127 |
262549_at |
AT1G31290
|
AGO3, ARGONAUTE 3 |
0.283 |
-0.052 |
| 128 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.283 |
-0.858 |
| 129 |
260668_at |
AT1G19530
|
unknown |
0.282 |
-0.246 |
| 130 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.282 |
0.053 |
| 131 |
260327_at |
AT1G63840
|
[RING/U-box superfamily protein] |
0.281 |
-0.092 |
| 132 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
0.281 |
-0.190 |
| 133 |
250098_at |
AT5G17350
|
unknown |
0.279 |
-0.715 |
| 134 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.278 |
-0.372 |
| 135 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.277 |
-0.934 |
| 136 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
0.276 |
-0.453 |
| 137 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
0.276 |
-0.675 |
| 138 |
247535_at |
AT5G61620
|
[myb-like transcription factor family protein] |
0.275 |
-0.103 |
| 139 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.274 |
-0.237 |
| 140 |
260744_at |
AT1G15010
|
unknown |
0.273 |
-0.737 |
| 141 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.273 |
-0.612 |
| 142 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.273 |
-0.567 |
| 143 |
254900_at |
AT4G11510
|
RALFL28, ralf-like 28 |
0.272 |
0.086 |
| 144 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.272 |
-0.502 |
| 145 |
253832_at |
AT4G27654
|
unknown |
0.271 |
-0.949 |
| 146 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.268 |
-0.225 |
| 147 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.264 |
-0.227 |
| 148 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
0.263 |
-0.084 |
| 149 |
249042_at |
AT5G44350
|
[ethylene-responsive nuclear protein -related] |
0.263 |
-0.124 |
| 150 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.263 |
-0.570 |
| 151 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
0.262 |
-0.040 |
| 152 |
264314_at |
AT1G70420
|
unknown |
0.261 |
0.306 |
| 153 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
0.260 |
-0.008 |
| 154 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.260 |
0.010 |
| 155 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.259 |
-0.661 |
| 156 |
247933_at |
AT5G56980
|
unknown |
0.259 |
-0.479 |
| 157 |
264289_at |
AT1G61890
|
[MATE efflux family protein] |
0.257 |
-0.345 |
| 158 |
256433_at |
AT3G10985
|
ATWI-12, ARABIDOPSIS THALIANA WOUND-INDUCED PROTEIN 12, SAG20, senescence associated gene 20, WI12, WOUND-INDUCED PROTEIN 12 |
0.256 |
-0.131 |
| 159 |
256177_at |
AT1G51620
|
[Protein kinase superfamily protein] |
0.254 |
-0.366 |
| 160 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.253 |
-0.289 |
| 161 |
247800_at |
AT5G58570
|
unknown |
0.253 |
0.422 |
| 162 |
255599_at |
AT4G01010
|
ATCNGC13, CYCLIC NUCLEOTIDE-GATED CHANNEL 13, CNGC13, cyclic nucleotide-gated channel 13 |
0.253 |
-0.306 |
| 163 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.252 |
-0.280 |
| 164 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
0.252 |
-0.171 |
| 165 |
259516_at |
AT1G20450
|
ERD10, EARLY RESPONSIVE TO DEHYDRATION 10, LTI29, LOW TEMPERATURE INDUCED 29, LTI45, LOW TEMPERATURE INDUCED 45 |
0.251 |
-0.122 |
| 166 |
252255_at |
AT3G49220
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.251 |
-0.085 |
| 167 |
245141_at |
AT2G45400
|
BEN1, BRI1-5 ENHANCED 1 |
0.250 |
-0.156 |
| 168 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.250 |
-0.847 |
| 169 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.249 |
-1.124 |
| 170 |
246584_at |
AT5G14730
|
unknown |
0.249 |
0.199 |
| 171 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.248 |
-0.076 |
| 172 |
254200_at |
AT4G24110
|
unknown |
0.248 |
-0.192 |
| 173 |
260023_at |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
0.248 |
0.007 |
| 174 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
0.245 |
-0.266 |
| 175 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.245 |
-0.519 |
| 176 |
256763_at |
AT3G16860
|
COBL8, COBRA-like protein 8 precursor |
0.243 |
-0.555 |
| 177 |
267518_at |
AT2G30500
|
NET4B, Networked 4B |
0.242 |
-0.133 |
| 178 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.241 |
-0.728 |
| 179 |
250289_at |
AT5G13190
|
AtGILP, GILP, GSH-induced LITAF domain protein |
0.240 |
-0.225 |
| 180 |
253276_at |
AT4G34050
|
CCoAOMT1, caffeoyl coenzyme A O-methyltransferase 1 |
0.239 |
-0.048 |
| 181 |
259976_at |
AT1G76560
|
CP12-3, CP12 domain-containing protein 3 |
0.239 |
-0.051 |
| 182 |
251984_at |
AT3G53260
|
ATPAL2, PAL2, phenylalanine ammonia-lyase 2 |
0.239 |
-0.126 |
| 183 |
262237_at |
AT1G48320
|
DHNAT1, DHNA-CoA thioesterase 1 |
0.238 |
-0.183 |
| 184 |
267209_at |
AT2G30930
|
unknown |
0.238 |
-0.141 |
| 185 |
260686_at |
AT1G17620
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.237 |
-0.212 |
| 186 |
264313_at |
AT1G70410
|
ATBCA4, BETA CARBONIC ANHYDRASE 4, BCA4, beta carbonic anhydrase 4, CA4, BETA CARBONIC ANHYDRASE 4 |
0.237 |
0.035 |
| 187 |
263931_at |
AT2G36220
|
unknown |
0.236 |
-0.273 |
| 188 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
0.235 |
0.023 |
| 189 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.234 |
-0.697 |
| 190 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.233 |
-0.308 |
| 191 |
248968_at |
AT5G45280
|
[Pectinacetylesterase family protein] |
0.233 |
-0.111 |
| 192 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
0.232 |
0.070 |
| 193 |
256840_x_at |
AT3G32000
|
[gypsy-like retrotransposon family (Athila), has a 5.4e-305 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-0.299 |
0.005 |
| 194 |
258303_at |
AT3G30620
|
[gypsy-like retrotransposon family (Athila)] |
-0.254 |
-0.016 |
| 195 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
-0.221 |
0.065 |
| 196 |
258231_at |
AT3G27730
|
MER3, RCK, ROCK-N-ROLLERS |
-0.214 |
-0.004 |
| 197 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.200 |
-0.010 |
| 198 |
259815_at |
AT1G49870
|
unknown |
-0.199 |
-0.011 |
| 199 |
256082_at |
AT1G20720
|
[RAD3-like DNA-binding helicase protein] |
-0.186 |
-0.047 |
| 200 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-0.182 |
0.260 |
| 201 |
250601_at |
AT5G07810
|
[SNF2 domain-containing protein / helicase domain-containing protein / HNH endonuclease domain-containing protein] |
-0.179 |
0.103 |
| 202 |
251527_at |
AT3G58650
|
TRM7, TON1 Recruiting Motif 7 |
-0.178 |
0.071 |
| 203 |
266439_s_at |
AT2G43200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.177 |
0.201 |
| 204 |
259003_at |
AT3G02010
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.173 |
-0.009 |
| 205 |
250269_at |
AT5G12970
|
[Calcium-dependent lipid-binding (CaLB domain) plant phosphoribosyltransferase family protein] |
-0.170 |
-0.015 |
| 206 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
-0.168 |
-0.015 |
| 207 |
255410_at |
AT4G03100
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
-0.167 |
0.197 |
| 208 |
262971_at |
AT1G75640
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.166 |
0.166 |
| 209 |
253053_at |
AT4G37470
|
KAI2, KARRIKIN INSENSITIVE 2 |
-0.165 |
0.042 |
| 210 |
260366_at |
AT1G70460
|
AtPERK13, proline-rich extensin-like receptor kinase 13, PERK13, proline-rich extensin-like receptor kinase 13, RHS10, root hair specific 10 |
-0.162 |
-0.097 |
| 211 |
246315_at |
AT3G56870
|
unknown |
-0.161 |
0.105 |
| 212 |
264599_at |
AT1G04600
|
ATXIA, XIA, myosin XI A |
-0.160 |
-0.080 |
| 213 |
247779_at |
AT5G58760
|
DDB2, damaged DNA binding 2 |
-0.159 |
0.071 |
| 214 |
256259_at |
AT3G12460
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.157 |
0.042 |
| 215 |
254288_at |
AT4G22970
|
AESP, homolog of separase, ESP, EXTRA SPINDLE POLES, ESP, homolog of separase, RSW4, RADIALLY SWOLLEN 4 |
-0.156 |
-0.065 |
| 216 |
252646_at |
AT3G44610
|
[Protein kinase superfamily protein] |
-0.155 |
-0.116 |
| 217 |
257201_at |
AT3G23740
|
unknown |
-0.154 |
0.276 |
| 218 |
247856_at |
AT5G58300
|
[Leucine-rich repeat protein kinase family protein] |
-0.153 |
-0.000 |
| 219 |
258741_at |
AT3G05790
|
LON4, lon protease 4 |
-0.150 |
-0.059 |
| 220 |
259780_at |
AT1G29630
|
[5'-3' exonuclease family protein] |
-0.150 |
0.051 |
| 221 |
258596_at |
AT3G04510
|
LSH2, LIGHT SENSITIVE HYPOCOTYLS 2 |
-0.149 |
-0.067 |
| 222 |
257542_at |
AT3G26050
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.149 |
0.082 |
| 223 |
247667_at |
AT5G60150
|
unknown |
-0.147 |
0.050 |
| 224 |
265964_at |
AT2G37510
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.145 |
0.013 |
| 225 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.145 |
-0.212 |
| 226 |
250386_at |
AT5G11510
|
AtMYB3R4, myb domain protein 3R4, MYB3R-4, myb domain protein 3r-4 |
-0.144 |
0.036 |
| 227 |
253251_at |
AT4G34730
|
RBF1, RbfA domain-containing protein 1 |
-0.143 |
-0.087 |
| 228 |
264604_at |
AT1G04650
|
unknown |
-0.142 |
0.004 |
| 229 |
251067_at |
AT5G01910
|
unknown |
-0.141 |
0.189 |
| 230 |
258867_at |
AT3G03130
|
unknown |
-0.141 |
0.110 |
| 231 |
259537_at |
AT1G12370
|
PHR1, photolyase 1, UVR2, UV RESISTANCE 2 |
-0.139 |
-0.127 |
| 232 |
262976_at |
AT1G75520
|
SRS5, SHI-related sequence 5 |
-0.139 |
-0.018 |
| 233 |
251727_at |
AT3G56290
|
unknown |
-0.139 |
-0.267 |
| 234 |
255613_at |
AT4G01270
|
[RING/U-box superfamily protein] |
-0.139 |
0.101 |
| 235 |
259222_at |
AT3G03680
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.138 |
0.066 |
| 236 |
254400_at |
AT4G21270
|
ATK1, kinesin 1, KATA, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA A, KATAP, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA A PROTEIN |
-0.138 |
-0.006 |
| 237 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.138 |
0.029 |
| 238 |
248057_at |
AT5G55520
|
unknown |
-0.136 |
0.128 |
| 239 |
263629_at |
AT2G04850
|
[Auxin-responsive family protein] |
-0.136 |
-0.087 |
| 240 |
254649_at |
AT4G18570
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.136 |
0.037 |
| 241 |
264251_at |
AT1G09190
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.136 |
0.006 |
| 242 |
253397_at |
AT4G32710
|
PERK14, proline-rich extensin-like receptor kinase 14 |
-0.135 |
0.047 |
| 243 |
259912_at |
AT1G72670
|
iqd8, IQ-domain 8 |
-0.134 |
0.107 |
| 244 |
247527_at |
AT5G61480
|
PXY, PHLOEM INTERCALATED WITH XYLEM, TDR, TDIF receptor |
-0.134 |
0.043 |
| 245 |
245453_at |
AT4G16900
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.131 |
-0.097 |
| 246 |
262758_at |
AT1G10780
|
[F-box/RNI-like superfamily protein] |
-0.131 |
0.160 |
| 247 |
265998_at |
AT2G24270
|
ALDH11A3, aldehyde dehydrogenase 11A3 |
-0.130 |
-0.048 |
| 248 |
255808_at |
AT4G10280
|
[RmlC-like cupins superfamily protein] |
-0.130 |
0.014 |
| 249 |
259433_at |
AT1G01570
|
[FUNCTIONS IN: transferase activity, transferring glycosyl groups] |
-0.130 |
0.222 |
| 250 |
255597_at |
AT4G01730
|
[DHHC-type zinc finger family protein] |
-0.129 |
0.196 |
| 251 |
253532_at |
AT4G31570
|
unknown |
-0.129 |
0.030 |
| 252 |
254173_at |
AT4G24580
|
REN1, ROP1 ENHANCER 1 |
-0.128 |
-0.006 |
| 253 |
255376_x_at |
AT4G03790
|
[gypsy-like retrotransposon family (Athila), has a 2.2e-286 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-0.128 |
0.034 |
| 254 |
263768_x_at |
AT2G06330
|
[pseudogene] |
-0.128 |
-0.081 |
| 255 |
260609_at |
AT2G43690
|
LecRK-V.3, L-type lectin receptor kinase V.3 |
-0.127 |
0.002 |
| 256 |
257740_at |
AT3G27330
|
[zinc finger (C3HC4-type RING finger) family protein] |
-0.127 |
0.134 |
| 257 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
-0.126 |
0.086 |
| 258 |
266508_at |
AT2G47920
|
NET3C, Networked 3C |
-0.126 |
0.037 |
| 259 |
253432_at |
AT4G32450
|
MEF8S, MEF8 similar |
-0.125 |
0.109 |
| 260 |
259151_at |
AT3G10310
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.125 |
0.120 |
| 261 |
260073_at |
AT1G73660
|
SIS8, SUGAR INSENSITIVE 8 |
-0.124 |
-0.108 |
| 262 |
247612_at |
AT5G60730
|
[Anion-transporting ATPase] |
-0.124 |
-0.002 |
| 263 |
251593_at |
AT3G57660
|
NRPA1, nuclear RNA polymerase A1 |
-0.123 |
0.001 |
| 264 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.123 |
-0.079 |
| 265 |
260667_at |
AT1G19440
|
KCS4, 3-ketoacyl-CoA synthase 4 |
-0.122 |
0.019 |
| 266 |
262462_at |
AT1G50350
|
unknown |
-0.122 |
0.063 |
| 267 |
258441_at |
AT3G17260
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 2.6e-27 P-value blast match to GB:CAA29005 ORFa of Maize Ac (hAT-element) (Zea mays)] |
-0.122 |
-0.035 |
| 268 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.121 |
-0.207 |
| 269 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.121 |
0.134 |
| 270 |
246137_at |
AT5G28490
|
LSH1, LIGHT-DEPENDENT SHORT HYPOCOTYLS 1, OBO2, ORGAN BOUNDARY 2 |
-0.121 |
0.076 |
| 271 |
249115_at |
AT5G43810
|
AGO10, ARGONAUTE 10, PNH, PINHEAD, ZLL, ZWILLE |
-0.119 |
0.203 |
| 272 |
245174_at |
AT2G47500
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.119 |
-0.082 |
| 273 |
263782_at |
AT2G46380
|
unknown |
-0.119 |
0.007 |
| 274 |
262337_at |
AT1G64260
|
[MuDR family transposase] |
-0.119 |
-0.013 |
| 275 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-0.118 |
0.019 |
| 276 |
246983_at |
AT5G67200
|
[Leucine-rich repeat protein kinase family protein] |
-0.118 |
0.072 |
| 277 |
260182_at |
AT1G70750
|
MyoB2, myosin binding protein 2 |
-0.117 |
0.099 |
| 278 |
253622_at |
AT4G30560
|
ATCNGC9, cyclic nucleotide gated channel 9, CNGC9, cyclic nucleotide gated channel 9 |
-0.117 |
0.274 |
| 279 |
248940_at |
AT5G45400
|
ATRPA70C, RPA70C |
-0.116 |
0.127 |
| 280 |
250756_at |
AT5G05940
|
ATROPGEF5, ROP GUANINE NUCLEOTIDE EXCHANGE FACTOR 5, ROPGEF5, ROP (rho of plants) guanine nucleotide exchange factor 5 |
-0.116 |
0.032 |
| 281 |
245259_at |
AT4G14150
|
KINESIN-12A, PAKRP1, phragmoplast-associated kinesin-related protein 1 |
-0.116 |
0.057 |
| 282 |
254933_at |
AT4G11130
|
RDR2, RNA-dependent RNA polymerase 2, SMD1, SILENCING MOVEMENT DEFICIENT 1 |
-0.116 |
0.070 |
| 283 |
253779_at |
AT4G28490
|
HAE, HAESA, RLK5, RECEPTOR-LIKE PROTEIN KINASE 5 |
-0.115 |
-0.042 |
| 284 |
260494_at |
AT2G41820
|
PXC3, PXY/TDR-correlated 3 |
-0.115 |
0.168 |
| 285 |
245403_at |
AT4G17590
|
unknown |
-0.115 |
0.024 |
| 286 |
247310_at |
AT5G63950
|
CHR24, chromatin remodeling 24 |
-0.115 |
0.022 |
| 287 |
248055_at |
AT5G55830
|
LecRK-S.7, L-type lectin receptor kinase S.7 |
-0.115 |
0.039 |
| 288 |
263913_at |
AT2G36570
|
PXC1, PXY/TDR-correlated 1 |
-0.115 |
-0.000 |
| 289 |
252025_at |
AT3G52900
|
unknown |
-0.114 |
0.162 |
| 290 |
249214_at |
AT5G42720
|
[Glycosyl hydrolase family 17 protein] |
-0.114 |
-0.065 |
| 291 |
259930_at |
AT1G34355
|
ATPS1, PARALLEL SPINDLE 1, PS1, PARALLEL SPINDLE 1 |
-0.114 |
0.030 |
| 292 |
259677_at |
AT1G77740
|
PIP5K2, phosphatidylinositol-4-phosphate 5-kinase 2 |
-0.113 |
-0.074 |
| 293 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.113 |
-0.015 |
| 294 |
257618_at |
AT3G24715
|
[Protein kinase superfamily protein with octicosapeptide/Phox/Bem1p domain] |
-0.113 |
-0.076 |
| 295 |
248420_at |
AT5G51560
|
[Leucine-rich repeat protein kinase family protein] |
-0.112 |
0.162 |
| 296 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
-0.112 |
-0.058 |
| 297 |
258628_at |
AT3G02890
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.112 |
-0.064 |
| 298 |
261961_at |
AT1G36510
|
[Nucleic acid-binding proteins superfamily] |
-0.111 |
-0.090 |
| 299 |
252355_at |
AT3G48250
|
BIR6, Buthionine sulfoximine-insensitive roots 6 |
-0.110 |
0.084 |
| 300 |
245739_at |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
-0.110 |
0.103 |
| 301 |
256623_at |
AT3G19960
|
ATM1, myosin 1 |
-0.110 |
0.152 |
| 302 |
252442_at |
AT3G46940
|
DUT1, DUTP-PYROPHOSPHATASE-LIKE 1 |
-0.110 |
0.031 |
| 303 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.110 |
0.132 |
| 304 |
256858_at |
AT3G15140
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.110 |
-0.007 |
| 305 |
246542_at |
AT5G15020
|
SNL2, SIN3-like 2 |
-0.109 |
-0.002 |
| 306 |
260684_at |
AT1G17590
|
NF-YA8, nuclear factor Y, subunit A8 |
-0.109 |
-0.125 |
| 307 |
255314_at |
AT4G04150
|
[gypsy-like retrotransposon family (Athila), has a 2.0e-27 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-0.109 |
0.001 |
| 308 |
250957_at |
AT5G03250
|
[Phototropic-responsive NPH3 family protein] |
-0.109 |
0.080 |
| 309 |
266009_at |
AT2G37420
|
[ATP binding microtubule motor family protein] |
-0.108 |
0.130 |
| 310 |
248603_at |
AT5G49430
|
[WD40/YVTN repeat-like-containing domain] |
-0.108 |
0.007 |
| 311 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.108 |
0.487 |
| 312 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
-0.108 |
-0.171 |
| 313 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
-0.107 |
-0.167 |
| 314 |
266479_at |
AT2G31160
|
LSH3, LIGHT SENSITIVE HYPOCOTYLS 3, OBO1, ORGAN BOUNDARY 1 |
-0.107 |
0.053 |
| 315 |
257364_at |
AT2G45940
|
unknown |
-0.107 |
0.002 |
| 316 |
262833_at |
AT1G14750
|
SDS, SOLO DANCERS |
-0.107 |
0.011 |
| 317 |
258914_at |
AT3G06360
|
AGP27, arabinogalactan protein 27, ATAGP27, ARABINOGALACTAN PROTEIN 27 |
-0.106 |
-0.006 |
| 318 |
264964_at |
AT1G60460
|
unknown |
-0.106 |
-0.038 |
| 319 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.106 |
-0.037 |
| 320 |
250103_at |
AT5G16600
|
AtMYB43, myb domain protein 43, MYB43, myb domain protein 43 |
-0.106 |
0.163 |
| 321 |
246415_at |
AT5G17160
|
unknown |
-0.106 |
0.312 |
| 322 |
246411_at |
AT1G57770
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.105 |
-0.102 |
| 323 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.105 |
0.170 |
| 324 |
258355_at |
AT3G14330
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.105 |
0.133 |
| 325 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.104 |
0.488 |
| 326 |
261085_at |
AT1G17480
|
IQD7, IQ-domain 7 |
-0.104 |
0.122 |
| 327 |
245607_at |
AT4G14330
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.104 |
0.100 |
| 328 |
246510_at |
AT5G15410
|
ATCNGC2, CYCLIC NUCLEOTIDE-GATED CHANNEL 2, CNGC2, CYCLIC NUCLEOTIDE GATED CHANNEL 2, DND1, DEFENSE NO DEATH 1 |
-0.103 |
-0.120 |
| 329 |
249354_at |
AT5G40480
|
EMB3012, embryo defective 3012 |
-0.103 |
-0.073 |
| 330 |
246765_at |
AT5G27330
|
[Prefoldin chaperone subunit family protein] |
-0.103 |
0.110 |
| 331 |
252272_at |
AT3G49670
|
BAM2, BARELY ANY MERISTEM 2 |
-0.101 |
0.097 |
| 332 |
248232_at |
AT5G53760
|
ATMLO11, MILDEW RESISTANCE LOCUS O 11, MLO11, MILDEW RESISTANCE LOCUS O 11 |
-0.101 |
0.073 |
| 333 |
247850_at |
AT5G58150
|
[Leucine-rich repeat protein kinase family protein] |
-0.101 |
-0.167 |
| 334 |
249989_at |
AT5G18525
|
[protein serine/threonine kinases] |
-0.101 |
0.238 |
| 335 |
251485_at |
AT3G59550
|
ATRAD21.2, SISTER CHROMATID COHESION 1 PROTEIN 3, ATSYN3, SYN3 |
-0.101 |
0.011 |
| 336 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
-0.101 |
-0.113 |
| 337 |
259128_at |
AT3G02260
|
ASA1, ATTENUATED SHADE AVOIDANCE 1, BIG, BIG, CRM1, CORYMBOSA1, DOC1, DARK OVER-EXPRESSION OF CAB 1, LPR1, LOW PHOSPHATE-RESISTANT ROOT 1, TIR3, TRANSPORT INHIBITOR RESPONSE 3, UMB1, UMBRELLA 1 |
-0.101 |
-0.005 |
| 338 |
258159_at |
AT3G17840
|
RLK902, receptor-like kinase 902 |
-0.100 |
-0.068 |
| 339 |
258965_at |
AT3G10530
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.100 |
0.131 |
| 340 |
244999_at |
ATCG00190
|
RPOB, RNA polymerase subunit beta |
-0.100 |
0.088 |
| 341 |
251868_at |
AT3G54490
|
RPB5E, RNA polymerase II fifth largest subunit, E |
-0.099 |
-0.069 |
| 342 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
-0.099 |
-0.986 |
| 343 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.099 |
0.055 |
| 344 |
249020_at |
AT5G44800
|
CHR4, chromatin remodeling 4, PKR1, PICKLE RELATED 1 |
-0.099 |
-0.000 |
| 345 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.099 |
0.020 |
| 346 |
266560_at |
AT2G23950
|
[Leucine-rich repeat protein kinase family protein] |
-0.099 |
0.219 |
| 347 |
247589_at |
AT5G60690
|
IFL, INTERFASCICULAR FIBERLESS, IFL1, INTERFASCICULAR FIBERLESS 1, REV, REVOLUTA |
-0.098 |
0.040 |
| 348 |
248653_at |
AT5G49290
|
ATRLP56, receptor like protein 56, RLP56, receptor like protein 56 |
-0.098 |
-0.019 |
| 349 |
256757_at |
AT3G25620
|
ABCG21, ATP-binding cassette G21 |
-0.098 |
-0.176 |
| 350 |
252207_at |
AT3G50370
|
unknown |
-0.098 |
0.012 |
| 351 |
266915_at |
AT2G45870
|
[Bestrophin-like protein] |
-0.098 |
0.058 |
| 352 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.098 |
-0.445 |
| 353 |
249426_at |
AT5G39840
|
[ATP-dependent RNA helicase, mitochondrial, putative] |
-0.098 |
0.094 |
| 354 |
258138_at |
AT3G24495
|
ATMSH7, ARABIDOPSIS THALIANA MUTS HOMOLOG 7, MSH6-2, MUTS HOMOLOG 6-2, MSH7, MUTS homolog 7 |
-0.097 |
0.067 |
| 355 |
260910_at |
AT1G02690
|
IMPA-6, importin alpha isoform 6 |
-0.097 |
0.014 |
| 356 |
257722_at |
AT3G18490
|
ASPG1, ASPARTIC PROTEASE IN GUARD CELL 1 |
-0.096 |
-0.082 |
| 357 |
252954_at |
AT4G38660
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.096 |
0.038 |
| 358 |
253368_at |
AT4G33200
|
ATXI-I, MYOSIN XI I, XI-15, MYOSIN XI-15, XI-I |
-0.096 |
-0.036 |
| 359 |
267406_at |
AT2G34780
|
EMB1611, EMBRYO DEFECTIVE 1611, MEE22, MATERNAL EFFECT EMBRYO ARREST 22 |
-0.096 |
0.060 |
| 360 |
253402_at |
AT4G32880
|
ATHB-8, homeobox gene 8, ATHB8, HB-8, homeobox gene 8 |
-0.096 |
-0.027 |
| 361 |
258462_at |
AT3G17350
|
unknown |
-0.095 |
0.051 |
| 362 |
247606_at |
AT5G61000
|
ATRPA70D, RPA70D |
-0.095 |
0.061 |
| 363 |
246817_at |
AT5G27240
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.095 |
0.081 |
| 364 |
258166_at |
AT3G21540
|
[transducin family protein / WD-40 repeat family protein] |
-0.095 |
-0.027 |
| 365 |
256341_at |
AT1G72040
|
AtdNK, dNK, deoxyribonucleoside kinase |
-0.095 |
-0.105 |
| 366 |
251865_at |
AT3G54930
|
[Protein phosphatase 2A regulatory B subunit family protein] |
-0.094 |
0.159 |
| 367 |
251955_at |
AT3G53680
|
[Acyl-CoA N-acyltransferase with RING/FYVE/PHD-type zinc finger domain] |
-0.094 |
-0.202 |
| 368 |
261290_at |
AT1G36990
|
unknown |
-0.093 |
-0.017 |
| 369 |
251942_at |
AT3G53480
|
ABCG37, ATP-binding cassette G37, ATPDR9, PLEIOTROPIC DRUG RESISTANCE 9, PDR9, pleiotropic drug resistance 9, PIS1, polar auxin transport inhibitor sensitive 1 |
-0.093 |
-0.009 |
| 370 |
266655_at |
AT2G25880
|
AtAUR2, ataurora2, AUR2, ataurora2 |
-0.093 |
0.080 |
| 371 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
-0.093 |
0.118 |
| 372 |
267402_at |
AT2G26180
|
IQD6, IQ-domain 6 |
-0.093 |
0.096 |
| 373 |
264452_at |
AT1G10270
|
GRP23, glutamine-rich protein 23 |
-0.093 |
-0.128 |
| 374 |
258502_at |
AT3G02490
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.092 |
-0.001 |
| 375 |
246010_at |
AT5G08440
|
unknown |
-0.092 |
0.029 |
| 376 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
-0.092 |
0.039 |
| 377 |
259444_at |
AT1G02370
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.091 |
0.038 |
| 378 |
255178_at |
AT4G08050
|
[gypsy-like retrotransposon family (Athila), has a 0. P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-0.091 |
0.138 |
| 379 |
264731_at |
AT1G62150
|
[Mitochondrial transcription termination factor family protein] |
-0.091 |
0.049 |
| 380 |
245210_at |
AT5G12350
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
-0.091 |
0.008 |
| 381 |
263605_at |
AT2G16485
|
NERD, Needed for RDR2-independent DNA methylation |
-0.091 |
0.138 |
| 382 |
256528_at |
AT1G66140
|
ZFP4, zinc finger protein 4 |
-0.091 |
0.146 |
| 383 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.091 |
-0.264 |