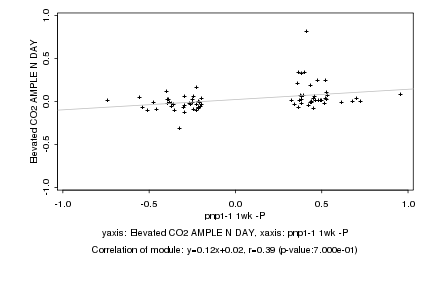
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pnp1-1 1wk -P |
Log2 signal ratio
Elevated CO2 AMPLE N DAY |
| 1 |
244977_at |
ATCG00730
|
PETD, photosynthetic electron transfer D |
0.954 |
0.083 |
| 2 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
0.724 |
0.010 |
| 3 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
0.698 |
0.046 |
| 4 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
0.675 |
0.003 |
| 5 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.611 |
-0.011 |
| 6 |
244966_at |
ATCG00600
|
PETG |
0.528 |
0.081 |
| 7 |
246888_at |
AT5G26270
|
unknown |
0.526 |
0.110 |
| 8 |
255893_at |
AT1G17960
|
[Threonyl-tRNA synthetase] |
0.522 |
0.026 |
| 9 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.520 |
0.249 |
| 10 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.516 |
0.041 |
| 11 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.514 |
-0.021 |
| 12 |
245015_at |
ATCG00490
|
RBCL |
0.498 |
0.020 |
| 13 |
265051_at |
AT1G52100
|
[Mannose-binding lectin superfamily protein] |
0.492 |
0.019 |
| 14 |
247024_at |
AT5G66985
|
unknown |
0.481 |
0.014 |
| 15 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.472 |
0.246 |
| 16 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.462 |
0.017 |
| 17 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.458 |
0.069 |
| 18 |
263515_at |
AT2G21640
|
unknown |
0.450 |
0.041 |
| 19 |
265428_at |
AT2G20720
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.448 |
-0.077 |
| 20 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.437 |
-0.010 |
| 21 |
245008_at |
ATCG00360
|
YCF3 |
0.436 |
0.010 |
| 22 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.434 |
0.191 |
| 23 |
245003_at |
ATCG00280
|
PSBC, photosystem II reaction center protein C |
0.431 |
-0.002 |
| 24 |
260522_x_at |
AT2G41730
|
unknown |
0.419 |
-0.037 |
| 25 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.407 |
0.818 |
| 26 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.399 |
0.349 |
| 27 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.392 |
0.071 |
| 28 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.382 |
0.328 |
| 29 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.380 |
-0.018 |
| 30 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.379 |
0.062 |
| 31 |
245047_at |
ATCG00020
|
PSBA, photosystem II reaction center protein A |
0.377 |
0.024 |
| 32 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.372 |
0.079 |
| 33 |
256188_at |
AT1G30160
|
unknown |
0.369 |
0.016 |
| 34 |
258203_at |
AT3G13950
|
unknown |
0.363 |
0.342 |
| 35 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.361 |
-0.067 |
| 36 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.355 |
0.220 |
| 37 |
245007_at |
ATCG00350
|
PSAA |
0.337 |
-0.033 |
| 38 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
0.321 |
0.017 |
| 39 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.747 |
0.023 |
| 40 |
264720_at |
AT1G70080
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.558 |
0.048 |
| 41 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
-0.542 |
-0.066 |
| 42 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.511 |
-0.104 |
| 43 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.476 |
-0.010 |
| 44 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
-0.461 |
-0.092 |
| 45 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.403 |
0.120 |
| 46 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.397 |
0.033 |
| 47 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.396 |
-0.014 |
| 48 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.393 |
0.032 |
| 49 |
259766_at |
AT1G64360
|
unknown |
-0.387 |
-0.007 |
| 50 |
252134_at |
AT3G50910
|
unknown |
-0.375 |
-0.013 |
| 51 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.372 |
-0.048 |
| 52 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.364 |
-0.031 |
| 53 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.356 |
-0.102 |
| 54 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.328 |
-0.303 |
| 55 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.304 |
-0.064 |
| 56 |
263544_at |
AT2G21590
|
APL4 |
-0.298 |
-0.039 |
| 57 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.297 |
-0.117 |
| 58 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
-0.296 |
0.065 |
| 59 |
251235_at |
AT3G62860
|
[alpha/beta-Hydrolases superfamily protein] |
-0.268 |
-0.012 |
| 60 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.264 |
-0.025 |
| 61 |
259371_at |
AT1G69080
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.252 |
-0.006 |
| 62 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.250 |
0.033 |
| 63 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.246 |
-0.087 |
| 64 |
252573_at |
AT3G45260
|
[C2H2-like zinc finger protein] |
-0.244 |
0.060 |
| 65 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.230 |
-0.098 |
| 66 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.229 |
-0.031 |
| 67 |
247469_at |
AT5G62165
|
AGL42, AGAMOUS-like 42, FYF, FOREVER YOUNG FLOWER |
-0.227 |
0.171 |
| 68 |
258633_at |
AT3G07990
|
SCPL27, serine carboxypeptidase-like 27 |
-0.219 |
0.007 |
| 69 |
255899_at |
AT1G17970
|
[RING/U-box superfamily protein] |
-0.217 |
-0.061 |
| 70 |
261134_at |
AT1G19630
|
CYP722A1, cytochrome P450, family 722, subfamily A, polypeptide 1 |
-0.215 |
-0.075 |
| 71 |
256920_at |
AT3G18980
|
ETP1, EIN2 targeting protein1 |
-0.215 |
-0.062 |
| 72 |
259340_at |
AT3G03870
|
unknown |
-0.213 |
-0.005 |
| 73 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.208 |
-0.057 |
| 74 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.202 |
0.038 |
| 75 |
263370_at |
AT2G20500
|
unknown |
-0.200 |
-0.025 |