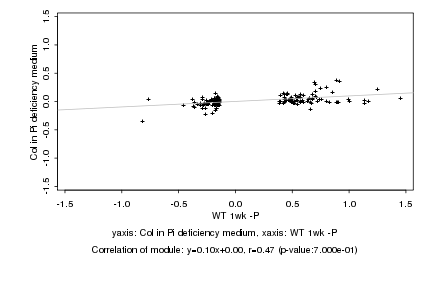
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT 1wk -P |
Log2 signal ratio
Col in Pi deficiency medium |
| 1 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
1.449 |
0.061 |
| 2 |
246001_at |
AT5G20790
|
unknown |
1.246 |
0.224 |
| 3 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
1.168 |
0.008 |
| 4 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
1.134 |
0.021 |
| 5 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
1.129 |
-0.025 |
| 6 |
266766_at |
AT2G46880
|
ATPAP14, PAP14, purple acid phosphatase 14 |
1.003 |
0.004 |
| 7 |
261814_at |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
0.991 |
0.047 |
| 8 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.908 |
0.366 |
| 9 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
0.903 |
-0.001 |
| 10 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.894 |
-0.013 |
| 11 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.885 |
0.374 |
| 12 |
250091_at |
AT5G17340
|
[Putative membrane lipoprotein] |
0.884 |
-0.011 |
| 13 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.850 |
0.164 |
| 14 |
253087_at |
AT4G36350
|
ATPAP25, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 25, PAP25, purple acid phosphatase 25 |
0.821 |
-0.016 |
| 15 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.795 |
0.260 |
| 16 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
0.793 |
0.004 |
| 17 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
0.755 |
0.036 |
| 18 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
0.740 |
0.241 |
| 19 |
252730_at |
AT3G43110
|
unknown |
0.736 |
0.044 |
| 20 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
0.717 |
0.005 |
| 21 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.712 |
0.082 |
| 22 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
0.704 |
0.193 |
| 23 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
0.702 |
0.304 |
| 24 |
254111_at |
AT4G24890
|
ATPAP24, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 24, PAP24, purple acid phosphatase 24 |
0.700 |
0.094 |
| 25 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.694 |
0.351 |
| 26 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.677 |
0.062 |
| 27 |
263851_at |
AT2G04460
|
unknown |
0.676 |
0.139 |
| 28 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
0.672 |
0.043 |
| 29 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
0.667 |
-0.028 |
| 30 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.659 |
-0.124 |
| 31 |
245882_at |
AT5G09470
|
DIC3, dicarboxylate carrier 3 |
0.657 |
-0.012 |
| 32 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
0.649 |
0.060 |
| 33 |
260201_at |
AT1G67600
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
0.648 |
0.067 |
| 34 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.639 |
0.039 |
| 35 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.629 |
0.013 |
| 36 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.596 |
0.110 |
| 37 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.593 |
-0.010 |
| 38 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.586 |
0.035 |
| 39 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
0.572 |
0.009 |
| 40 |
252006_at |
AT3G52820
|
ATPAP22, PURPLE ACID PHOSPHATASE 22, PAP22, purple acid phosphatase 22 |
0.567 |
0.127 |
| 41 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
0.567 |
0.077 |
| 42 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.565 |
-0.015 |
| 43 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.561 |
0.072 |
| 44 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.554 |
0.085 |
| 45 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.546 |
-0.042 |
| 46 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.544 |
0.074 |
| 47 |
256014_at |
AT1G19200
|
unknown |
0.539 |
0.068 |
| 48 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.539 |
0.028 |
| 49 |
249465_at |
AT5G39720
|
AIG2L, avirulence induced gene 2 like protein |
0.532 |
0.056 |
| 50 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.531 |
0.020 |
| 51 |
247393_at |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.523 |
0.112 |
| 52 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
0.517 |
0.017 |
| 53 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.513 |
-0.025 |
| 54 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.506 |
-0.033 |
| 55 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.504 |
0.000 |
| 56 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.502 |
0.003 |
| 57 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.493 |
0.076 |
| 58 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.490 |
0.006 |
| 59 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.485 |
0.039 |
| 60 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.485 |
-0.007 |
| 61 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.480 |
0.061 |
| 62 |
267646_at |
AT2G32830
|
PHT1;5, phosphate transporter 1;5, PHT5, PHOSPHATE TRANSPORTER 5 |
0.467 |
0.019 |
| 63 |
254290_at |
AT4G23000
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
0.466 |
0.021 |
| 64 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
0.453 |
0.156 |
| 65 |
258682_at |
AT3G08720
|
ATPK19, Arabidopsis thaliana protein kinase 19, ATPK2, ARABIDOPSIS THALIANA PROTEIN KINASE 2, ATS6K2, ARABIDOPSIS THALIANA SERINE/THREONINE PROTEIN KINASE 2, S6K2, serine/threonine protein kinase 2 |
0.452 |
0.127 |
| 66 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.448 |
0.131 |
| 67 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
0.448 |
0.021 |
| 68 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
0.446 |
0.027 |
| 69 |
260254_at |
AT1G74210
|
AtGDPD5, GDPD5, glycerophosphodiester phosphodiesterase 5 |
0.436 |
0.036 |
| 70 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.429 |
0.128 |
| 71 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.428 |
0.073 |
| 72 |
258293_at |
AT3G23430
|
ATPHO1, ARABIDOPSIS PHOSPHATE 1, PHO1, phosphate 1 |
0.427 |
0.014 |
| 73 |
265334_at |
AT2G18370
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.427 |
-0.006 |
| 74 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.427 |
0.041 |
| 75 |
253044_at |
AT4G37290
|
unknown |
0.421 |
0.144 |
| 76 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
0.417 |
-0.023 |
| 77 |
256627_at |
AT3G19970
|
[alpha/beta-Hydrolases superfamily protein] |
0.394 |
0.114 |
| 78 |
258182_at |
AT3G21500
|
DXL1, DXS-like 1, DXPS1, 1-deoxy-D-xylulose 5-phosphate synthase 1, DXS2, 1-deoxy-D-xylulose 5-phosphate (DXP) synthase 2 |
0.390 |
0.004 |
| 79 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
0.381 |
0.011 |
| 80 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
0.379 |
-0.018 |
| 81 |
261684_at |
AT1G47400
|
unknown |
-0.822 |
-0.341 |
| 82 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.773 |
0.053 |
| 83 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.464 |
-0.055 |
| 84 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.383 |
0.048 |
| 85 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.372 |
-0.074 |
| 86 |
266616_at |
AT2G29680
|
ATCDC6, CDC6, cell division control 6 |
-0.367 |
-0.094 |
| 87 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
-0.365 |
-0.007 |
| 88 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.335 |
-0.036 |
| 89 |
256569_at |
AT3G19550
|
unknown |
-0.312 |
-0.024 |
| 90 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
-0.312 |
-0.057 |
| 91 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.298 |
0.075 |
| 92 |
251704_at |
AT3G56360
|
unknown |
-0.293 |
-0.118 |
| 93 |
267580_at |
AT2G41990
|
unknown |
-0.292 |
0.036 |
| 94 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.288 |
-0.067 |
| 95 |
262464_at |
AT1G50280
|
[Phototropic-responsive NPH3 family protein] |
-0.285 |
-0.042 |
| 96 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.275 |
-0.024 |
| 97 |
262238_at |
AT1G48300
|
DGAT3, diacylglycerol acyltransferase 3 |
-0.264 |
-0.123 |
| 98 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.264 |
-0.217 |
| 99 |
254954_at |
AT4G10910
|
unknown |
-0.262 |
0.024 |
| 100 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.255 |
-0.040 |
| 101 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.249 |
-0.035 |
| 102 |
259996_at |
AT1G67910
|
unknown |
-0.247 |
-0.003 |
| 103 |
264638_at |
AT1G65480
|
FT, FLOWERING LOCUS T, RSB8, REDUCED STEM BRANCHING 8 |
-0.242 |
-0.005 |
| 104 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.240 |
-0.001 |
| 105 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-0.238 |
-0.044 |
| 106 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.234 |
-0.014 |
| 107 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.231 |
0.012 |
| 108 |
261308_at |
AT1G48480
|
RKL1, receptor-like kinase 1 |
-0.227 |
0.018 |
| 109 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.219 |
0.048 |
| 110 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.216 |
0.029 |
| 111 |
248432_at |
AT5G51390
|
unknown |
-0.210 |
-0.068 |
| 112 |
244939_at |
ATCG00065
|
RPS12, RIBOSOMAL PROTEIN S12, RPS12A, ribosomal protein S12A |
-0.209 |
0.033 |
| 113 |
245296_at |
AT4G16370
|
oligopeptide transporter |
-0.205 |
-0.206 |
| 114 |
248924_at |
AT5G45960
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.197 |
0.001 |
| 115 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.190 |
-0.060 |
| 116 |
245828_at |
AT1G57820
|
ORTH2, ORTHRUS 2, VIM1, VARIANT IN METHYLATION 1 |
-0.190 |
-0.025 |
| 117 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.189 |
-0.052 |
| 118 |
261638_at |
AT1G49975
|
[INVOLVED IN: photosynthesis] |
-0.189 |
0.032 |
| 119 |
248279_at |
AT5G52910
|
ATIM, TIMELESS |
-0.189 |
-0.028 |
| 120 |
252946_at |
AT4G39235
|
unknown |
-0.188 |
0.067 |
| 121 |
261388_at |
AT1G05385
|
LPA19, LOW PSII ACCUMULATION 19, Psb27-H1 |
-0.185 |
0.057 |
| 122 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
-0.185 |
-0.047 |
| 123 |
253010_at |
AT4G37750
|
ANT, AINTEGUMENTA, CKC, CKC1, COMPLEMENTING A PROTEIN KINASE C MUTANT 1, DRG, DRAGON |
-0.184 |
0.046 |
| 124 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
-0.184 |
0.146 |
| 125 |
261765_at |
AT1G15570
|
CYCA2;3, CYCLIN A2;3 |
-0.181 |
-0.004 |
| 126 |
266335_at |
AT2G32440
|
ATKAO2, ARABIDOPSIS ENT-KAURENOIC ACID HYDROXYLASE 2, CYP88A4, KAO2, ent-kaurenoic acid hydroxylase 2 |
-0.181 |
-0.064 |
| 127 |
255829_at |
AT2G40540
|
ATKT2, ATKUP2, KT2, potassium transporter 2, KUP2, SHY3, TRK2 |
-0.179 |
0.037 |
| 128 |
267578_at |
AT2G30695
|
unknown |
-0.177 |
-0.014 |
| 129 |
252501_at |
AT3G46880
|
unknown |
-0.176 |
-0.150 |
| 130 |
253495_at |
AT4G31850
|
PGR3, proton gradient regulation 3 |
-0.172 |
0.057 |
| 131 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.172 |
-0.119 |
| 132 |
250223_at |
AT5G14070
|
ROXY2 |
-0.171 |
-0.110 |
| 133 |
249861_at |
AT5G22875
|
unknown |
-0.171 |
0.038 |
| 134 |
258355_at |
AT3G14330
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.171 |
-0.033 |
| 135 |
260082_at |
AT1G78180
|
[Mitochondrial substrate carrier family protein] |
-0.169 |
0.017 |
| 136 |
253589_at |
AT4G30825
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.169 |
-0.015 |
| 137 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
-0.168 |
-0.013 |
| 138 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
-0.167 |
-0.005 |
| 139 |
258421_at |
AT3G16690
|
AtSWEET16, SWEET16 |
-0.167 |
-0.039 |
| 140 |
265349_at |
AT2G22610
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.167 |
-0.003 |
| 141 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.165 |
-0.063 |
| 142 |
250453_at |
AT5G10620
|
[methyltransferases] |
-0.164 |
-0.022 |
| 143 |
248977_at |
AT5G45020
|
[Glutathione S-transferase family protein] |
-0.161 |
0.005 |
| 144 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
-0.159 |
0.050 |
| 145 |
259160_at |
AT3G05410
|
[Photosystem II reaction center PsbP family protein] |
-0.158 |
0.094 |
| 146 |
246792_at |
AT5G27290
|
unknown |
-0.158 |
-0.033 |
| 147 |
249392_at |
AT5G40150
|
[Peroxidase superfamily protein] |
-0.158 |
0.016 |
| 148 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.158 |
-0.004 |
| 149 |
256970_at |
AT3G21090
|
ABCG15, ATP-binding cassette G15 |
-0.157 |
-0.031 |
| 150 |
246818_at |
AT5G27270
|
EMB976, EMBRYO DEFECTIVE 976 |
-0.155 |
-0.048 |
| 151 |
249129_at |
AT5G43080
|
CYCA3;1, Cyclin A3;1 |
-0.155 |
-0.055 |
| 152 |
259209_at |
AT3G09160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.154 |
0.079 |
| 153 |
266589_at |
AT2G46250
|
[myosin heavy chain-related] |
-0.154 |
0.064 |
| 154 |
246885_at |
AT5G26230
|
MAKR1, membrane-associated kinase regulator 1 |
-0.152 |
-0.042 |
| 155 |
260685_at |
AT1G17650
|
AtGLYR2, GLYR2, glyoxylate reductase 2, GR2, GLYOXYLATE REDUCTASE 2 |
-0.149 |
0.039 |
| 156 |
255709_at |
AT4G00180
|
YAB3, YABBY3 |
-0.147 |
0.027 |
| 157 |
251976_at |
AT3G53240
|
AtRLP45, receptor like protein 45, RLP45, receptor like protein 45 |
-0.146 |
-0.067 |
| 158 |
247826_at |
AT5G58480
|
[O-Glycosyl hydrolases family 17 protein] |
-0.146 |
0.000 |
| 159 |
249210_at |
AT5G42670
|
[Agenet domain-containing protein] |
-0.146 |
0.014 |