|
probeID |
AGICode |
Annotation |
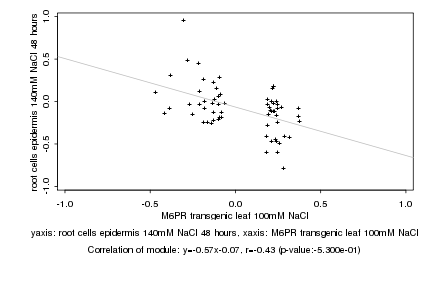
Log2 signal ratio
M6PR transgenic leaf 100mM NaCl |
Log2 signal ratio
root cells epidermis 140mM NaCl 48 hours |
| 1 |
259996_at |
AT1G67910
|
unknown |
0.375 |
-0.234 |
| 2 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.369 |
-0.168 |
| 3 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
0.365 |
-0.078 |
| 4 |
258468_at |
AT3G06070
|
unknown |
0.313 |
-0.419 |
| 5 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.286 |
-0.408 |
| 6 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
0.281 |
-0.779 |
| 7 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.265 |
-0.064 |
| 8 |
265275_at |
AT2G28440
|
[proline-rich family protein] |
0.256 |
-0.492 |
| 9 |
259530_at |
AT1G12450
|
[SNARE associated Golgi protein family] |
0.246 |
-0.240 |
| 10 |
263937_at |
AT2G35910
|
[RING/U-box superfamily protein] |
0.242 |
-0.027 |
| 11 |
264770_at |
AT1G23030
|
[ARM repeat superfamily protein] |
0.241 |
-0.597 |
| 12 |
254671_at |
AT4G18410
|
[Mutator-like transposase family, has a 8.7e-30 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.241 |
-0.079 |
| 13 |
250683_x_at |
AT5G06640
|
EXT10, extensin 10 |
0.240 |
-0.468 |
| 14 |
263177_at |
AT1G05540
|
unknown |
0.236 |
-0.155 |
| 15 |
258032_at |
AT3G21170
|
[F-box and associated interaction domains-containing protein] |
0.235 |
0.006 |
| 16 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
0.232 |
-0.443 |
| 17 |
252240_at |
AT3G50010
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.228 |
-0.117 |
| 18 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
0.223 |
-0.109 |
| 19 |
251231_at |
AT3G62760
|
ATGSTF13 |
0.221 |
0.178 |
| 20 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
0.221 |
0.179 |
| 21 |
262585_at |
AT1G15460
|
ATBOR4, ARABIDOPSIS THALIANA REQUIRES HIGH BORON 4, BOR4, REQUIRES HIGH BORON 4 |
0.219 |
-0.018 |
| 22 |
262760_at |
AT1G10770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.215 |
0.158 |
| 23 |
261594_at |
AT1G33240
|
AT-GTL1, GT2-LIKE 1, ATGTL1, GTL1, GT-2-like 1 |
0.211 |
-0.462 |
| 24 |
267281_at |
AT2G19400
|
[AGC (cAMP-dependent, cGMP-dependent and protein kinase C) kinase family protein] |
0.211 |
-0.109 |
| 25 |
252788_at |
AT3G42690
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT3G42690.1)] |
0.210 |
0.006 |
| 26 |
263527_at |
AT2G24810
|
[Pathogenesis-related thaumatin superfamily protein] |
0.202 |
-0.100 |
| 27 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.198 |
-0.063 |
| 28 |
255835_at |
AT2G33420
|
unknown |
0.190 |
-0.147 |
| 29 |
253136_at |
AT4G35470
|
PIRL4, plant intracellular ras group-related LRR 4 |
0.188 |
-0.272 |
| 30 |
250180_at |
AT5G14450
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.185 |
0.026 |
| 31 |
258808_at |
AT3G04050
|
[Pyruvate kinase family protein] |
0.184 |
-0.035 |
| 32 |
261675_at |
AT1G18290
|
unknown |
0.180 |
-0.404 |
| 33 |
254324_at |
AT4G22640
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.178 |
-0.593 |
| 34 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
-0.470 |
0.110 |
| 35 |
249813_at |
AT5G23940
|
DCR, DEFECTIVE IN CUTICULAR RIDGES, EMB3009, EMBRYO DEFECTIVE 3009, PEL3, PERMEABLE LEAVES3 |
-0.421 |
-0.131 |
| 36 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
-0.387 |
-0.072 |
| 37 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.384 |
0.308 |
| 38 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
-0.310 |
0.966 |
| 39 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
-0.285 |
0.493 |
| 40 |
263972_at |
AT2G42760
|
unknown |
-0.272 |
-0.033 |
| 41 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
-0.256 |
-0.148 |
| 42 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
-0.218 |
0.450 |
| 43 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
-0.212 |
0.120 |
| 44 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
-0.211 |
-0.031 |
| 45 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
-0.193 |
0.268 |
| 46 |
257951_at |
AT3G21700
|
ATSGP2, SGP2 |
-0.192 |
-0.239 |
| 47 |
264426_at |
AT1G61760
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.187 |
0.010 |
| 48 |
267144_at |
AT2G38110
|
ATGPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6, GPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6 |
-0.187 |
-0.078 |
| 49 |
248176_at |
AT5G54650
|
ATFH5, FORMIN HOMOLOGY 5, Fh5, formin homology5 |
-0.164 |
-0.245 |
| 50 |
260227_at |
AT1G74450
|
unknown |
-0.145 |
-0.251 |
| 51 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.140 |
-0.022 |
| 52 |
247913_at |
AT5G57510
|
unknown |
-0.130 |
-0.219 |
| 53 |
257968_at |
AT3G27550
|
[RNA-binding CRS1 / YhbY (CRM) domain protein] |
-0.129 |
0.232 |
| 54 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
-0.129 |
-0.125 |
| 55 |
265184_at |
AT1G23710
|
unknown |
-0.123 |
0.027 |
| 56 |
253219_at |
AT4G34990
|
AtMYB32, myb domain protein 32, MYB32, myb domain protein 32 |
-0.113 |
0.156 |
| 57 |
249833_at |
AT5G23430
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.105 |
-0.201 |
| 58 |
257014_at |
AT3G26930
|
[Protein with RNI-like/FBD-like domains] |
-0.103 |
0.064 |
| 59 |
259884_at |
AT1G76390
|
PUB43, plant U-box 43 |
-0.101 |
-0.031 |
| 60 |
254304_at |
AT4G22270
|
ATMRB1, ARABIDOPSIS MEMBRANE RELATED BIGGER1, MRB1, MEMBRANE RELATED BIGGER1 |
-0.098 |
-0.187 |
| 61 |
254761_at |
AT4G13195
|
CLE44, CLAVATA3/ESR-RELATED 44 |
-0.094 |
0.289 |
| 62 |
262249_at |
AT1G48380
|
HYP7, HYPOCOTYL 7, RHL1, ROOT HAIRLESS 1 |
-0.088 |
0.092 |
| 63 |
265886_at |
AT2G25620
|
AtDBP1, DNA-binding protein phosphatase 1, DBP1, DNA-binding protein phosphatase 1 |
-0.086 |
-0.180 |
| 64 |
247429_at |
AT5G62620
|
[Galactosyltransferase family protein] |
-0.083 |
-0.123 |
| 65 |
255594_at |
AT4G01660
|
ABC1, ABC transporter 1, ATABC1, ABC transporter 1, ATATH10 |
-0.065 |
-0.015 |