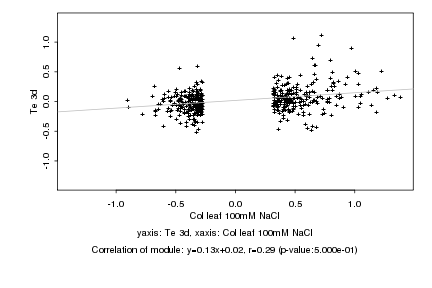
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col leaf 100mM NaCl |
Log2 signal ratio
Te 3d |
| 1 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.380 |
0.083 |
| 2 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.327 |
0.115 |
| 3 |
267459_at |
AT2G33850
|
unknown |
1.266 |
0.061 |
| 4 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
1.223 |
0.515 |
| 5 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
1.187 |
0.156 |
| 6 |
256442_at |
AT3G10930
|
unknown |
1.180 |
-0.182 |
| 7 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
1.179 |
0.227 |
| 8 |
264580_at |
AT1G05340
|
unknown |
1.151 |
0.199 |
| 9 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
1.136 |
-0.056 |
| 10 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
1.111 |
0.162 |
| 11 |
266658_at |
AT2G25735
|
unknown |
1.050 |
0.125 |
| 12 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
1.044 |
-0.029 |
| 13 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
1.043 |
0.090 |
| 14 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
1.029 |
0.475 |
| 15 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
1.025 |
0.296 |
| 16 |
263182_at |
AT1G05575
|
unknown |
1.023 |
-0.100 |
| 17 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
1.005 |
0.091 |
| 18 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
1.000 |
0.515 |
| 19 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.968 |
0.906 |
| 20 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.939 |
0.406 |
| 21 |
258225_at |
AT3G15630
|
unknown |
0.918 |
0.287 |
| 22 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.898 |
-0.089 |
| 23 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
0.885 |
0.077 |
| 24 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.870 |
0.127 |
| 25 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.866 |
0.064 |
| 26 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.860 |
0.346 |
| 27 |
261088_at |
AT1G07590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.846 |
0.091 |
| 28 |
253643_at |
AT4G29780
|
unknown |
0.846 |
-0.053 |
| 29 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.823 |
0.266 |
| 30 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.820 |
0.332 |
| 31 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.816 |
0.320 |
| 32 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.811 |
0.498 |
| 33 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.810 |
0.201 |
| 34 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.804 |
0.256 |
| 35 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.802 |
-0.223 |
| 36 |
260744_at |
AT1G15010
|
unknown |
0.793 |
0.694 |
| 37 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.790 |
0.404 |
| 38 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.785 |
-0.005 |
| 39 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.781 |
0.057 |
| 40 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.771 |
0.197 |
| 41 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.771 |
-0.015 |
| 42 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.770 |
0.035 |
| 43 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.766 |
0.017 |
| 44 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.764 |
0.187 |
| 45 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.743 |
-0.199 |
| 46 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.732 |
-0.122 |
| 47 |
246929_at |
AT5G25210
|
unknown |
0.728 |
0.066 |
| 48 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.725 |
-0.215 |
| 49 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.719 |
1.127 |
| 50 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.705 |
0.089 |
| 51 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.693 |
0.956 |
| 52 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.687 |
-0.031 |
| 53 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.680 |
0.082 |
| 54 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.676 |
0.373 |
| 55 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.674 |
0.011 |
| 56 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.673 |
-0.426 |
| 57 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.668 |
0.620 |
| 58 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
0.661 |
0.031 |
| 59 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.661 |
0.619 |
| 60 |
246200_at |
AT4G37240
|
unknown |
0.659 |
0.160 |
| 61 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.656 |
0.469 |
| 62 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
0.652 |
0.034 |
| 63 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.649 |
0.332 |
| 64 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.643 |
-0.009 |
| 65 |
262211_at |
AT1G74930
|
ORA47 |
0.643 |
-0.405 |
| 66 |
247177_at |
AT5G65300
|
unknown |
0.640 |
0.184 |
| 67 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.639 |
0.736 |
| 68 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
0.633 |
0.293 |
| 69 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.631 |
-0.475 |
| 70 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.621 |
-0.011 |
| 71 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
0.620 |
-0.214 |
| 72 |
266119_at |
AT2G02100
|
LCR69, low-molecular-weight cysteine-rich 69, PDF2.2 |
0.619 |
0.154 |
| 73 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
0.615 |
0.080 |
| 74 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
0.615 |
0.106 |
| 75 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
0.608 |
-0.060 |
| 76 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
0.597 |
0.109 |
| 77 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.597 |
0.263 |
| 78 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.597 |
-0.443 |
| 79 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.595 |
0.101 |
| 80 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.594 |
0.152 |
| 81 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
0.588 |
0.040 |
| 82 |
259979_at |
AT1G76600
|
unknown |
0.586 |
-0.042 |
| 83 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.581 |
-0.377 |
| 84 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
0.571 |
0.130 |
| 85 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
0.569 |
0.011 |
| 86 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.567 |
0.003 |
| 87 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.565 |
-0.234 |
| 88 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.564 |
-0.052 |
| 89 |
256675_at |
AT3G52170
|
[DNA binding] |
0.562 |
-0.175 |
| 90 |
264704_at |
AT1G70090
|
GATL9, GALACTURONOSYLTRANSFERASE-LIKE 9, LGT8, glucosyl transferase family 8 |
0.551 |
0.037 |
| 91 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
0.550 |
0.010 |
| 92 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.549 |
-0.109 |
| 93 |
261193_at |
AT1G32920
|
unknown |
0.548 |
0.009 |
| 94 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.544 |
0.058 |
| 95 |
255733_at |
AT1G25400
|
unknown |
0.541 |
0.187 |
| 96 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.539 |
0.451 |
| 97 |
253078_at |
AT4G36180
|
[Leucine-rich receptor-like protein kinase family protein] |
0.537 |
-0.059 |
| 98 |
259303_at |
AT3G05130
|
unknown |
0.535 |
-0.003 |
| 99 |
265276_at |
AT2G28400
|
unknown |
0.528 |
0.069 |
| 100 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
0.527 |
-0.217 |
| 101 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.524 |
0.081 |
| 102 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.520 |
0.058 |
| 103 |
260656_at |
AT1G19380
|
unknown |
0.518 |
0.292 |
| 104 |
262452_at |
AT1G11210
|
unknown |
0.511 |
0.131 |
| 105 |
253292_at |
AT4G33985
|
unknown |
0.508 |
0.242 |
| 106 |
267280_at |
AT2G19450
|
ABX45, AS11, ATDGAT, Arabidopsis thaliana acyl-CoA:diacylglycerol acyltransferase, AtDGAT1, DGAT1, acyl-CoA:diacylglycerol acyltransferase 1, RDS1, TAG1, TRIACYLGLYCEROL BIOSYNTHESIS DEFECT 1 |
0.507 |
0.157 |
| 107 |
247937_at |
AT5G57110
|
ACA8, autoinhibited Ca2+ -ATPase, isoform 8, AT-ACA8, AUTOINHIBITED CA2+ -ATPASE, ISOFORM 8 |
0.504 |
0.217 |
| 108 |
246253_at |
AT4G37260
|
ATMYB73, MYB73, myb domain protein 73 |
0.501 |
-0.013 |
| 109 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.496 |
-0.069 |
| 110 |
245789_at |
AT1G32090
|
[early-responsive to dehydration stress protein (ERD4)] |
0.490 |
0.052 |
| 111 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.483 |
1.064 |
| 112 |
258505_at |
AT3G06530
|
[ARM repeat superfamily protein] |
0.483 |
0.145 |
| 113 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.482 |
-0.131 |
| 114 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.482 |
0.239 |
| 115 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.481 |
-0.073 |
| 116 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.481 |
0.062 |
| 117 |
249977_at |
AT5G18820
|
Cpn60alpha2, chaperonin-60alpha2, EMB3007, embryo defective 3007 |
0.481 |
-0.026 |
| 118 |
247708_at |
AT5G59550
|
unknown |
0.479 |
-0.105 |
| 119 |
257667_at |
AT3G20440
|
BE1, BRANCHING ENZYME 1, EMB2729, EMBRYO DEFECTIVE 2729 |
0.478 |
-0.012 |
| 120 |
249872_at |
AT5G23130
|
[Peptidoglycan-binding LysM domain-containing protein] |
0.473 |
-0.161 |
| 121 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
0.467 |
0.187 |
| 122 |
246289_at |
AT3G56880
|
[VQ motif-containing protein] |
0.467 |
0.029 |
| 123 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
0.463 |
0.135 |
| 124 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.462 |
0.194 |
| 125 |
256890_at |
AT3G23830
|
AtGRP4, GR-RBP4, GRP4, glycine-rich RNA-binding protein 4 |
0.460 |
-0.001 |
| 126 |
248247_at |
AT5G53210
|
SPCH, SPEECHLESS |
0.456 |
0.037 |
| 127 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.454 |
-0.167 |
| 128 |
256858_at |
AT3G15140
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.453 |
-0.023 |
| 129 |
251355_at |
AT3G61100
|
[Putative endonuclease or glycosyl hydrolase] |
0.453 |
0.052 |
| 130 |
251610_at |
AT3G57930
|
unknown |
0.453 |
0.049 |
| 131 |
250787_at |
AT5G05560
|
APC1, anaphase promoting complex 1, EMB2771, EMBRYO DEFECTIVE 2771 |
0.453 |
-0.024 |
| 132 |
265184_at |
AT1G23710
|
unknown |
0.453 |
-0.183 |
| 133 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.450 |
0.148 |
| 134 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.448 |
0.060 |
| 135 |
256284_at |
AT3G12530
|
PSF2 |
0.448 |
-0.157 |
| 136 |
247279_at |
AT5G64310
|
AGP1, arabinogalactan protein 1, ATAGP1 |
0.446 |
0.174 |
| 137 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.445 |
0.411 |
| 138 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.445 |
0.214 |
| 139 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.443 |
-0.121 |
| 140 |
247410_at |
AT5G62990
|
emb1692, embryo defective 1692 |
0.443 |
0.013 |
| 141 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.443 |
0.310 |
| 142 |
261727_at |
AT1G76090
|
SMT3, sterol methyltransferase 3 |
0.442 |
0.132 |
| 143 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.441 |
0.022 |
| 144 |
257193_at |
AT3G13160
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.436 |
0.033 |
| 145 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
0.435 |
0.196 |
| 146 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
0.435 |
0.308 |
| 147 |
267349_at |
AT2G40010
|
[Ribosomal protein L10 family protein] |
0.433 |
0.045 |
| 148 |
246018_at |
AT5G10695
|
unknown |
0.433 |
-0.025 |
| 149 |
247240_at |
AT5G64660
|
ATCMPG2, CMPG2, CYS, MET, PRO, and GLY protein 2 |
0.433 |
0.150 |
| 150 |
245041_at |
AT2G26530
|
AR781 |
0.431 |
0.056 |
| 151 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.431 |
0.194 |
| 152 |
245699_at |
AT5G04250
|
[Cysteine proteinases superfamily protein] |
0.431 |
0.403 |
| 153 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.429 |
-0.317 |
| 154 |
261984_at |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
0.429 |
-0.056 |
| 155 |
258254_at |
AT3G26782
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.427 |
0.088 |
| 156 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
0.425 |
0.003 |
| 157 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
0.425 |
-0.068 |
| 158 |
265539_at |
AT2G15830
|
unknown |
0.425 |
0.301 |
| 159 |
251192_at |
AT3G62720
|
ATXT1, XT1, xylosyltransferase 1, XXT1, XYG XYLOSYLTRANSFERASE 1 |
0.420 |
0.133 |
| 160 |
267623_at |
AT2G39650
|
unknown |
0.417 |
-0.149 |
| 161 |
265326_at |
AT2G18220
|
[Noc2p family] |
0.414 |
0.111 |
| 162 |
267529_at |
AT2G45490
|
AtAUR3, ataurora3, AUR3, ataurora3 |
0.414 |
0.045 |
| 163 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
0.411 |
-0.015 |
| 164 |
260841_at |
AT1G29195
|
unknown |
0.408 |
-0.049 |
| 165 |
253981_at |
AT4G26670
|
[Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein] |
0.407 |
0.036 |
| 166 |
265732_at |
AT2G01300
|
unknown |
0.404 |
0.280 |
| 167 |
253733_at |
AT4G29170
|
ATMND1 |
0.403 |
-0.089 |
| 168 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.402 |
-0.122 |
| 169 |
261272_at |
AT1G26665
|
[Mediator complex, subunit Med10] |
0.400 |
-0.091 |
| 170 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
0.398 |
0.051 |
| 171 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.398 |
-0.284 |
| 172 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.395 |
-0.219 |
| 173 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
0.394 |
0.084 |
| 174 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.392 |
0.272 |
| 175 |
259479_at |
AT1G19020
|
unknown |
0.392 |
0.087 |
| 176 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
0.389 |
-0.160 |
| 177 |
253614_at |
AT4G30350
|
SMXL2, SMAX1-like 2 |
0.387 |
0.271 |
| 178 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.386 |
0.149 |
| 179 |
248100_at |
AT5G55180
|
[O-Glycosyl hydrolases family 17 protein] |
0.384 |
0.428 |
| 180 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.381 |
0.187 |
| 181 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
0.379 |
0.026 |
| 182 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
0.376 |
0.170 |
| 183 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.375 |
0.193 |
| 184 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
0.374 |
-0.014 |
| 185 |
250679_at |
AT5G06550
|
JMJ22, Jumonji domain-containing protein 22 |
0.373 |
0.080 |
| 186 |
264624_at |
AT1G08930
|
ERD6, EARLY RESPONSE TO DEHYDRATION 6 |
0.372 |
0.103 |
| 187 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
0.371 |
-0.046 |
| 188 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.370 |
-0.329 |
| 189 |
261124_at |
AT1G04900
|
unknown |
0.364 |
0.028 |
| 190 |
254208_at |
AT4G24175
|
unknown |
0.364 |
0.055 |
| 191 |
249940_at |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
0.362 |
-0.197 |
| 192 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.362 |
0.011 |
| 193 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.360 |
-0.140 |
| 194 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
0.358 |
0.081 |
| 195 |
255844_at |
AT2G33580
|
LYK5, LysM-containing receptor-like kinase 5 |
0.357 |
-0.467 |
| 196 |
258468_at |
AT3G06070
|
unknown |
0.357 |
0.354 |
| 197 |
257395_at |
AT2G15630
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.356 |
0.020 |
| 198 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.355 |
-0.073 |
| 199 |
260126_at |
AT1G36370
|
SHM7, serine hydroxymethyltransferase 7 |
0.355 |
0.012 |
| 200 |
259071_at |
AT3G11650
|
NHL2, NDR1/HIN1-like 2 |
0.351 |
-0.098 |
| 201 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.351 |
0.445 |
| 202 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
0.351 |
0.102 |
| 203 |
256755_at |
AT3G25600
|
[Calcium-binding EF-hand family protein] |
0.350 |
-0.140 |
| 204 |
246898_at |
AT5G25580
|
unknown |
0.349 |
-0.078 |
| 205 |
251640_at |
AT3G57450
|
unknown |
0.349 |
-0.040 |
| 206 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.348 |
0.112 |
| 207 |
251800_at |
AT3G55510
|
RBL, REBELOTE |
0.347 |
0.072 |
| 208 |
264512_at |
AT1G09575
|
unknown |
0.345 |
0.248 |
| 209 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
0.345 |
0.051 |
| 210 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
0.342 |
0.309 |
| 211 |
267069_at |
AT2G41010
|
ATCAMBP25, calmodulin (CAM)-binding protein of 25 kDa, CAMBP25, calmodulin (CAM)-binding protein of 25 kDa |
0.338 |
-0.063 |
| 212 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
0.336 |
-0.035 |
| 213 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
0.336 |
-0.002 |
| 214 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
0.334 |
-0.006 |
| 215 |
252770_at |
AT3G42860
|
[zinc knuckle (CCHC-type) family protein] |
0.331 |
0.155 |
| 216 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.327 |
0.081 |
| 217 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
0.327 |
0.033 |
| 218 |
253175_at |
AT4G35050
|
MSI3, MULTICOPY SUPPRESSOR OF IRA1 3, NFC3, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 3 |
0.327 |
-0.026 |
| 219 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.326 |
0.123 |
| 220 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.326 |
0.192 |
| 221 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
0.324 |
0.419 |
| 222 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
0.321 |
-0.120 |
| 223 |
256525_at |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
0.320 |
0.212 |
| 224 |
260227_at |
AT1G74450
|
unknown |
0.320 |
-0.174 |
| 225 |
257524_at |
AT3G01330
|
DEL3, DP-E2F-like protein 3, E2FF, E2L2, E2F-LIKE 2 |
0.320 |
0.031 |
| 226 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
0.319 |
0.118 |
| 227 |
263864_at |
AT2G04530
|
CPZ, TRZ2, TRNASE Z 2 |
0.318 |
-0.032 |
| 228 |
247280_at |
AT5G64260
|
EXL2, EXORDIUM like 2 |
0.317 |
0.139 |
| 229 |
267393_at |
AT2G44500
|
[O-fucosyltransferase family protein] |
0.316 |
-0.074 |
| 230 |
255856_at |
AT1G66940
|
[protein kinase-related] |
0.316 |
0.123 |
| 231 |
260206_at |
AT1G70740
|
[Protein kinase superfamily protein] |
0.316 |
-0.075 |
| 232 |
252483_at |
AT3G46600
|
[GRAS family transcription factor] |
0.316 |
-0.061 |
| 233 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.314 |
0.233 |
| 234 |
263221_at |
AT1G30620
|
HSR8, HIGH SUGAR RESPONSE8, MUR4, MURUS 4, UXE1, UDP-D-XYLOSE 4-EPIMERASE 1 |
0.313 |
0.172 |
| 235 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.912 |
0.031 |
| 236 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.903 |
-0.086 |
| 237 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.779 |
-0.210 |
| 238 |
247055_at |
AT5G66740
|
unknown |
-0.696 |
0.087 |
| 239 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.685 |
0.259 |
| 240 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
-0.676 |
-0.150 |
| 241 |
263545_at |
AT2G21560
|
unknown |
-0.676 |
-0.155 |
| 242 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-0.672 |
-0.232 |
| 243 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.653 |
-0.044 |
| 244 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
-0.652 |
-0.121 |
| 245 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
-0.633 |
-0.038 |
| 246 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
-0.617 |
0.050 |
| 247 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.606 |
-0.413 |
| 248 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-0.604 |
0.003 |
| 249 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
-0.599 |
0.120 |
| 250 |
261247_at |
AT1G20070
|
unknown |
-0.599 |
0.054 |
| 251 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
-0.570 |
-0.153 |
| 252 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.568 |
-0.114 |
| 253 |
252619_at |
AT3G45210
|
unknown |
-0.567 |
0.084 |
| 254 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.565 |
0.176 |
| 255 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.551 |
-0.165 |
| 256 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
-0.551 |
0.003 |
| 257 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.548 |
0.021 |
| 258 |
254145_at |
AT4G24700
|
unknown |
-0.545 |
-0.245 |
| 259 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.541 |
-0.138 |
| 260 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.539 |
0.068 |
| 261 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.530 |
-0.056 |
| 262 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.516 |
0.097 |
| 263 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.511 |
-0.018 |
| 264 |
257057_at |
AT3G15310
|
unknown |
-0.510 |
0.168 |
| 265 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.500 |
-0.051 |
| 266 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
-0.497 |
0.206 |
| 267 |
255377_at |
AT4G03500
|
[Ankyrin repeat family protein] |
-0.497 |
0.089 |
| 268 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.496 |
-0.288 |
| 269 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.487 |
0.120 |
| 270 |
257451_at |
AT1G05690
|
BT3, BTB and TAZ domain protein 3 |
-0.487 |
-0.148 |
| 271 |
249759_at |
AT5G24380
|
ATYSL2, YELLOW STRIPE LIKE 2, YSL2, YELLOW STRIPE like 2 |
-0.484 |
0.010 |
| 272 |
261845_at |
AT1G15960
|
ATNRAMP6, NRAMP6, NRAMP metal ion transporter 6 |
-0.481 |
0.030 |
| 273 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.479 |
-0.102 |
| 274 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.476 |
-0.038 |
| 275 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.473 |
-0.235 |
| 276 |
250327_at |
AT5G12050
|
unknown |
-0.470 |
0.563 |
| 277 |
261448_at |
AT1G21140
|
[Vacuolar iron transporter (VIT) family protein] |
-0.470 |
-0.052 |
| 278 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
-0.470 |
0.049 |
| 279 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
-0.469 |
-0.064 |
| 280 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
-0.468 |
0.038 |
| 281 |
250589_at |
AT5G07700
|
AtMYB76, myb domain protein 76, MYB76, myb domain protein 76 |
-0.467 |
0.063 |
| 282 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-0.466 |
-0.362 |
| 283 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.464 |
-0.112 |
| 284 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
-0.463 |
-0.166 |
| 285 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
-0.461 |
0.058 |
| 286 |
245090_at |
AT2G40900
|
UMAMIT11, Usually multiple acids move in and out Transporters 11 |
-0.460 |
0.112 |
| 287 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
-0.460 |
-0.067 |
| 288 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
-0.458 |
0.043 |
| 289 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
-0.456 |
0.163 |
| 290 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-0.455 |
0.003 |
| 291 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.448 |
-0.099 |
| 292 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.447 |
-0.008 |
| 293 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
-0.438 |
-0.050 |
| 294 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
-0.434 |
-0.170 |
| 295 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
-0.434 |
0.089 |
| 296 |
264479_at |
AT1G77280
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.429 |
0.175 |
| 297 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.429 |
-0.197 |
| 298 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
-0.428 |
-0.133 |
| 299 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.426 |
-0.015 |
| 300 |
261048_at |
AT1G01420
|
UGT72B3, UDP-glucosyl transferase 72B3 |
-0.422 |
0.184 |
| 301 |
246142_at |
AT5G19970
|
unknown |
-0.420 |
0.085 |
| 302 |
256927_at |
AT3G22550
|
unknown |
-0.417 |
0.100 |
| 303 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.415 |
-0.074 |
| 304 |
260491_at |
AT1G51440
|
DALL2, DAD1-Like Lipase 2 |
-0.414 |
-0.183 |
| 305 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.412 |
-0.338 |
| 306 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.411 |
-0.419 |
| 307 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.399 |
0.065 |
| 308 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.399 |
-0.121 |
| 309 |
267503_at |
AT2G45600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.398 |
-0.244 |
| 310 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.397 |
0.014 |
| 311 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.396 |
0.086 |
| 312 |
255406_at |
AT4G03450
|
[Ankyrin repeat family protein] |
-0.396 |
-0.158 |
| 313 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
-0.396 |
-0.191 |
| 314 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
-0.396 |
0.069 |
| 315 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.394 |
-0.160 |
| 316 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.393 |
0.228 |
| 317 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
-0.393 |
-0.299 |
| 318 |
245384_at |
AT4G16790
|
[hydroxyproline-rich glycoprotein family protein] |
-0.391 |
0.168 |
| 319 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.391 |
-0.046 |
| 320 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.390 |
0.123 |
| 321 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.389 |
-0.135 |
| 322 |
251058_at |
AT5G01790
|
unknown |
-0.388 |
-0.053 |
| 323 |
261772_at |
AT1G76240
|
unknown |
-0.387 |
0.063 |
| 324 |
256293_at |
AT1G69440
|
AGO7, ARGONAUTE7, ZIP, ZIPPY |
-0.384 |
-0.088 |
| 325 |
265012_at |
AT1G24470
|
ATKCR2, KCR2, beta-ketoacyl reductase 2 |
-0.383 |
0.034 |
| 326 |
260601_at |
AT1G55910
|
ZIP11, zinc transporter 11 precursor |
-0.382 |
-0.231 |
| 327 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
-0.378 |
-0.017 |
| 328 |
262736_at |
AT1G28570
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.376 |
-0.145 |
| 329 |
263115_at |
AT1G03055
|
AtD27, A. thaliana homolog of rice D27, D27, DWARF27 |
-0.375 |
0.041 |
| 330 |
260696_at |
AT1G32520
|
unknown |
-0.374 |
-0.147 |
| 331 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.373 |
0.096 |
| 332 |
266363_at |
AT2G41250
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.372 |
-0.114 |
| 333 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.372 |
0.110 |
| 334 |
264130_at |
AT1G79160
|
unknown |
-0.367 |
0.188 |
| 335 |
258038_at |
AT3G21260
|
GLTP3, GLYCOLIPID TRANSFER PROTEIN 3 |
-0.366 |
-0.126 |
| 336 |
261073_at |
AT1G07300
|
[josephin protein-related] |
-0.365 |
0.049 |
| 337 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.365 |
0.127 |
| 338 |
250396_at |
AT5G10970
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.364 |
-0.026 |
| 339 |
257456_at |
AT2G18120
|
SRS4, SHI-related sequence 4 |
-0.364 |
-0.010 |
| 340 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
-0.362 |
-0.033 |
| 341 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.361 |
-0.381 |
| 342 |
260259_at |
AT1G74300
|
[alpha/beta-Hydrolases superfamily protein] |
-0.358 |
-0.221 |
| 343 |
267413_at |
AT2G34960
|
CAT5, cationic amino acid transporter 5 |
-0.356 |
0.085 |
| 344 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.355 |
0.044 |
| 345 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
-0.355 |
-0.080 |
| 346 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-0.355 |
0.100 |
| 347 |
264958_at |
AT1G76960
|
unknown |
-0.355 |
-0.134 |
| 348 |
261425_at |
AT1G18880
|
AtNPF2.9, NPF2.9, NRT1/ PTR family 2.9, NRT1.9, nitrate transporter 1.9 |
-0.353 |
-0.302 |
| 349 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
-0.353 |
0.065 |
| 350 |
257858_at |
AT3G12920
|
BRG3, BOI-related gene 3 |
-0.352 |
-0.119 |
| 351 |
246523_at |
AT5G15850
|
ATCOL1, BBX2, B-box domain protein 2, COL1, CONSTANS-like 1 |
-0.352 |
0.182 |
| 352 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.351 |
-0.186 |
| 353 |
256066_at |
AT1G06980
|
unknown |
-0.351 |
0.268 |
| 354 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
-0.347 |
-0.298 |
| 355 |
249542_at |
AT5G38140
|
NF-YC12, nuclear factor Y, subunit C12 |
-0.346 |
-0.036 |
| 356 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
-0.345 |
-0.187 |
| 357 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.344 |
-0.397 |
| 358 |
248039_at |
AT5G55950
|
[Nucleotide/sugar transporter family protein] |
-0.344 |
0.035 |
| 359 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.343 |
-0.084 |
| 360 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
-0.342 |
-0.185 |
| 361 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
-0.342 |
-0.026 |
| 362 |
264609_at |
AT1G04530
|
TPR4, tetratricopeptide repeat 4 |
-0.341 |
-0.092 |
| 363 |
254175_at |
AT4G24050
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.341 |
0.064 |
| 364 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
-0.340 |
-0.327 |
| 365 |
264738_at |
AT1G62250
|
unknown |
-0.339 |
-0.088 |
| 366 |
259209_at |
AT3G09160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.339 |
0.037 |
| 367 |
267063_at |
AT2G41120
|
unknown |
-0.338 |
-0.352 |
| 368 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.337 |
-0.071 |
| 369 |
253665_at |
AT4G30230
|
unknown |
-0.334 |
0.083 |
| 370 |
250803_at |
AT5G04980
|
[DNAse I-like superfamily protein] |
-0.332 |
0.328 |
| 371 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.331 |
0.037 |
| 372 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
-0.331 |
0.207 |
| 373 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
-0.331 |
-0.147 |
| 374 |
251906_at |
AT3G53720
|
ATCHX20, cation/H+ exchanger 20, CHX20, cation/H+ exchanger 20 |
-0.330 |
0.035 |
| 375 |
255794_at |
AT2G33480
|
ANAC041, NAC domain containing protein 41, NAC041, NAC domain containing protein 41 |
-0.329 |
0.114 |
| 376 |
260466_at |
AT1G10900
|
[Phosphatidylinositol-4-phosphate 5-kinase family protein] |
-0.329 |
-0.099 |
| 377 |
257986_at |
AT3G20865
|
AGP40, arabinogalactan protein 40 |
-0.328 |
0.060 |
| 378 |
252661_at |
AT3G44450
|
unknown |
-0.328 |
0.119 |
| 379 |
252010_at |
AT3G52740
|
unknown |
-0.328 |
0.058 |
| 380 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.328 |
0.045 |
| 381 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.327 |
0.043 |
| 382 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.327 |
-0.511 |
| 383 |
251725_at |
AT3G56260
|
unknown |
-0.326 |
0.044 |
| 384 |
245561_at |
AT4G15500
|
UGT84A4 |
-0.326 |
0.109 |
| 385 |
247596_at |
AT5G60840
|
unknown |
-0.326 |
-0.129 |
| 386 |
246125_at |
AT5G19875
|
unknown |
-0.326 |
0.049 |
| 387 |
254301_at |
AT4G22790
|
[MATE efflux family protein] |
-0.325 |
-0.061 |
| 388 |
252073_at |
AT3G51750
|
unknown |
-0.325 |
-0.174 |
| 389 |
255627_at |
AT4G00955
|
unknown |
-0.324 |
-0.067 |
| 390 |
267535_at |
AT2G41940
|
ZFP8, zinc finger protein 8 |
-0.323 |
-0.047 |
| 391 |
267070_at |
AT2G41000
|
[Chaperone DnaJ-domain superfamily protein] |
-0.322 |
0.068 |
| 392 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.322 |
-0.219 |
| 393 |
259179_at |
AT3G01660
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.321 |
0.051 |
| 394 |
258919_at |
AT3G10525
|
LGO, LOSS OF GIANT CELLS FROM ORGANS, SMR1, SIAMESE RELATED 1 |
-0.321 |
0.171 |
| 395 |
247601_at |
AT5G60850
|
OBP4, OBF binding protein 4 |
-0.320 |
0.069 |
| 396 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.320 |
0.154 |
| 397 |
248801_at |
AT5G47370
|
HAT2 |
-0.320 |
0.600 |
| 398 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
-0.319 |
0.298 |
| 399 |
248440_at |
AT5G51260
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.318 |
0.128 |
| 400 |
263875_at |
AT2G21970
|
SEP2, stress enhanced protein 2 |
-0.318 |
-0.337 |
| 401 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
-0.316 |
-0.459 |
| 402 |
250683_x_at |
AT5G06640
|
EXT10, extensin 10 |
-0.316 |
0.136 |
| 403 |
249112_at |
AT5G43780
|
APS4 |
-0.315 |
0.025 |
| 404 |
251107_at |
AT5G01610
|
unknown |
-0.315 |
-0.072 |
| 405 |
258925_at |
AT3G10420
|
SPD1, SEEDLING PLASTID DEVELOPMENT 1 |
-0.315 |
-0.121 |
| 406 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.315 |
-0.207 |
| 407 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
-0.314 |
-0.072 |
| 408 |
265146_at |
AT1G51210
|
[UDP-Glycosyltransferase superfamily protein] |
-0.313 |
0.058 |
| 409 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
-0.313 |
0.073 |
| 410 |
264310_at |
AT1G62030
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.312 |
-0.098 |
| 411 |
246462_at |
AT5G16940
|
[carbon-sulfur lyases] |
-0.312 |
-0.171 |
| 412 |
261056_at |
AT1G01360
|
PYL9, PYRABACTIN RESISTANCE 1-LIKE 9, RCAR1, regulatory component of ABA receptor 1 |
-0.307 |
-0.079 |
| 413 |
263311_at |
AT2G10340
|
unknown |
-0.306 |
0.029 |
| 414 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
-0.305 |
-0.009 |
| 415 |
260254_at |
AT1G74210
|
AtGDPD5, GDPD5, glycerophosphodiester phosphodiesterase 5 |
-0.305 |
-0.091 |
| 416 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.304 |
-0.044 |
| 417 |
246894_at |
AT5G25530
|
[DNAJ heat shock family protein] |
-0.303 |
0.021 |
| 418 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.303 |
-0.113 |
| 419 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.302 |
-0.159 |
| 420 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.301 |
0.051 |
| 421 |
267077_at |
AT2G40970
|
MYBC1 |
-0.301 |
-0.088 |
| 422 |
267096_at |
AT2G38180
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.300 |
-0.233 |
| 423 |
264843_at |
AT1G03400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.300 |
-0.151 |
| 424 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.299 |
-0.133 |
| 425 |
263693_at |
AT1G31200
|
ATPP2-A9, phloem protein 2-A9, PP2-A9, phloem protein 2-A9 |
-0.299 |
0.113 |
| 426 |
259299_at |
AT3G05080
|
unknown |
-0.299 |
0.056 |
| 427 |
260277_at |
AT1G80520
|
[Sterile alpha motif (SAM) domain-containing protein] |
-0.298 |
-0.059 |
| 428 |
256819_at |
AT3G21390
|
[Mitochondrial substrate carrier family protein] |
-0.298 |
-0.018 |
| 429 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
-0.298 |
-0.105 |
| 430 |
249134_at |
AT5G43150
|
unknown |
-0.297 |
-0.026 |
| 431 |
258905_at |
AT3G06390
|
[Uncharacterised protein family (UPF0497)] |
-0.297 |
0.117 |
| 432 |
250167_at |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-0.296 |
-0.019 |
| 433 |
245152_at |
AT2G47490
|
ATNDT1, ARABIDOPSIS THALIANA NAD+ TRANSPORTER 1, NDT1, NAD+ transporter 1 |
-0.296 |
0.044 |
| 434 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.295 |
0.001 |
| 435 |
261139_at |
AT1G19700
|
BEL10, BEL1-like homeodomain 10, BLH10, BEL1-LIKE HOMEODOMAIN 10 |
-0.295 |
-0.208 |
| 436 |
253100_at |
AT4G37400
|
CYP81F3, cytochrome P450, family 81, subfamily F, polypeptide 3 |
-0.293 |
0.187 |
| 437 |
260167_at |
AT1G71970
|
unknown |
-0.293 |
0.078 |
| 438 |
258049_at |
AT3G16220
|
[Predicted eukaryotic LigT] |
-0.293 |
-0.058 |
| 439 |
257610_at |
AT3G13810
|
AtIDD11, indeterminate(ID)-domain 11, IDD11, indeterminate(ID)-domain 11 |
-0.293 |
0.204 |
| 440 |
260309_at |
AT1G70580
|
AOAT2, alanine-2-oxoglutarate aminotransferase 2, GGT2, GLUTAMATE:GLYOXYLATE AMINOTRANSFERASE 2 |
-0.292 |
-0.118 |
| 441 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
-0.291 |
-0.012 |
| 442 |
265399_at |
AT2G11010
|
unknown |
-0.291 |
0.086 |
| 443 |
267066_at |
AT2G41040
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.291 |
0.006 |
| 444 |
266235_at |
AT2G02360
|
AtPP2-B10, phloem protein 2-B10, PP2-B10, phloem protein 2-B10 |
-0.291 |
0.045 |
| 445 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
-0.291 |
-0.057 |
| 446 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.290 |
0.121 |
| 447 |
251733_at |
AT3G56240
|
CCH, copper chaperone |
-0.290 |
-0.152 |
| 448 |
257519_at |
AT3G01210
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.288 |
0.017 |
| 449 |
253401_at |
AT4G32870
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.288 |
-0.112 |
| 450 |
245302_at |
AT4G17695
|
KAN3, KANADI 3 |
-0.288 |
-0.092 |
| 451 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.287 |
-0.182 |
| 452 |
255912_at |
AT1G66960
|
LUP5 |
-0.287 |
-0.034 |
| 453 |
245416_at |
AT4G17350
|
unknown |
-0.286 |
-0.062 |
| 454 |
254281_at |
AT4G22840
|
[Sodium Bile acid symporter family] |
-0.286 |
-0.101 |
| 455 |
250400_at |
AT5G10740
|
[Protein phosphatase 2C family protein] |
-0.286 |
0.143 |
| 456 |
261456_at |
AT1G21050
|
unknown |
-0.286 |
0.351 |
| 457 |
256926_at |
AT3G22540
|
unknown |
-0.286 |
0.014 |
| 458 |
252951_at |
AT4G38700
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.285 |
-0.023 |
| 459 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
-0.284 |
0.333 |
| 460 |
265990_at |
AT2G24280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.284 |
-0.126 |
| 461 |
261158_at |
AT1G34500
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.283 |
0.047 |
| 462 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.283 |
-0.074 |
| 463 |
261294_at |
AT1G48430
|
[Dihydroxyacetone kinase] |
-0.282 |
-0.213 |
| 464 |
252421_at |
AT3G47540
|
[Chitinase family protein] |
-0.281 |
-0.025 |
| 465 |
255944_at |
AT5G28600
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT1G35770.1)] |
-0.281 |
0.105 |
| 466 |
249932_at |
AT5G22390
|
unknown |
-0.281 |
-0.338 |
| 467 |
265128_at |
AT1G30860
|
[RING/U-box superfamily protein] |
-0.281 |
0.107 |