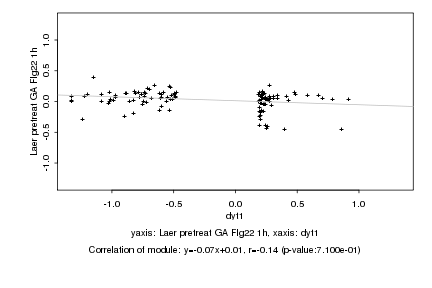
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
dyt1 |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
266524_at |
AT2G16960
|
[ARM repeat superfamily protein] |
0.909 |
0.041 |
| 2 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.854 |
-0.450 |
| 3 |
262010_at |
AT1G35612
|
[expressed protein] |
0.784 |
0.044 |
| 4 |
255414_at |
AT4G03156
|
[small GTPase-related] |
0.697 |
0.058 |
| 5 |
256987_at |
AT3G28560
|
[BCS1 AAA-type ATPase] |
0.666 |
0.102 |
| 6 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
0.580 |
0.068 |
| 7 |
260355_at |
AT1G69180
|
CRC, CRABS CLAW |
0.484 |
0.122 |
| 8 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
0.478 |
0.156 |
| 9 |
264826_at |
AT1G03410
|
2A6 |
0.429 |
0.017 |
| 10 |
259656_at |
AT1G55200
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.413 |
0.093 |
| 11 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
0.392 |
-0.449 |
| 12 |
251680_at |
AT3G57060
|
[binding] |
0.337 |
0.064 |
| 13 |
265086_at |
AT1G03770
|
ATRING1B, ARABIDOPSIS THALIANA RING 1B, RING1B, RING 1B |
0.334 |
0.107 |
| 14 |
263808_at |
AT2G04340
|
unknown |
0.302 |
0.060 |
| 15 |
250078_at |
AT5G16690
|
ATORC3, ORC3, origin recognition complex subunit 3 |
0.300 |
0.094 |
| 16 |
253859_at |
AT4G27657
|
unknown |
0.286 |
-0.050 |
| 17 |
256701_at |
AT3G52270
|
[Transcription initiation factor IIF, beta subunit] |
0.283 |
0.065 |
| 18 |
255904_at |
AT1G17860
|
[Kunitz family trypsin and protease inhibitor protein] |
0.278 |
0.058 |
| 19 |
264139_at |
AT1G78940
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.273 |
0.084 |
| 20 |
259151_at |
AT3G10310
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
0.270 |
0.065 |
| 21 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
0.269 |
0.009 |
| 22 |
266959_at |
AT2G07690
|
MCM5, MINICHROMOSOME MAINTENANCE 5 |
0.268 |
0.263 |
| 23 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
0.264 |
0.033 |
| 24 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
0.256 |
-0.406 |
| 25 |
245813_at |
AT1G49920
|
[MuDR family transposase] |
0.256 |
0.038 |
| 26 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
0.246 |
-0.434 |
| 27 |
266423_at |
AT2G41340
|
RPB5D, RNA polymerase II fifth largest subunit, D |
0.245 |
0.036 |
| 28 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
0.243 |
-0.041 |
| 29 |
263782_at |
AT2G46380
|
unknown |
0.242 |
0.048 |
| 30 |
258113_at |
AT3G14650
|
CYP72A11, cytochrome P450, family 72, subfamily A, polypeptide 11 |
0.241 |
-0.389 |
| 31 |
252128_at |
AT3G50870
|
GATA18, GATA TRANSCRIPTION FACTOR 18, HAN, HANABA TANARU, MNP, MONOPOLE |
0.239 |
0.067 |
| 32 |
245174_at |
AT2G47500
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
0.234 |
-0.037 |
| 33 |
249225_at |
AT5G42140
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
0.232 |
0.134 |
| 34 |
264074_at |
AT2G10780
|
[gypsy-like retrotransposon family, has a 0. P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
0.231 |
0.050 |
| 35 |
254177_at |
AT4G23860
|
[PHD finger protein-related] |
0.226 |
0.144 |
| 36 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
0.225 |
-0.041 |
| 37 |
248133_at |
AT5G54850
|
unknown |
0.222 |
0.129 |
| 38 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
0.221 |
-0.162 |
| 39 |
252129_at |
AT3G50890
|
AtHB28, homeobox protein 28, HB28, homeobox protein 28, ZHD7, ZINC FINGER HOMEODOMAIN 7 |
0.215 |
0.066 |
| 40 |
265348_at |
AT2G22600
|
[RNA-binding KH domain-containing protein] |
0.212 |
0.174 |
| 41 |
259252_at |
AT3G07610
|
IBM1, increase in bonsai methylation 1 |
0.209 |
0.104 |
| 42 |
256293_at |
AT1G69440
|
AGO7, ARGONAUTE7, ZIP, ZIPPY |
0.208 |
-0.017 |
| 43 |
262127_at |
AT1G52550
|
unknown |
0.208 |
-0.141 |
| 44 |
255770_at |
AT1G18560
|
[BED zinc finger ] |
0.207 |
0.130 |
| 45 |
264884_at |
AT1G61170
|
unknown |
0.204 |
0.034 |
| 46 |
264838_at |
AT1G03430
|
AHP5, histidine-containing phosphotransfer factor 5 |
0.202 |
-0.020 |
| 47 |
245604_at |
AT4G14290
|
[alpha/beta-Hydrolases superfamily protein] |
0.201 |
0.090 |
| 48 |
261605_at |
AT1G49580
|
[Calcium-dependent protein kinase (CDPK) family protein] |
0.200 |
-0.177 |
| 49 |
263199_at |
AT1G05590
|
ATHEX3, BETA-HEXOSAMINIDASE 3, HEXO2, beta-hexosaminidase 2 |
0.199 |
-0.281 |
| 50 |
254982_at |
AT4G10470
|
unknown |
0.198 |
-0.226 |
| 51 |
260544_at |
AT2G43540
|
unknown |
0.195 |
0.093 |
| 52 |
262840_at |
AT1G14900
|
HMGA, high mobility group A |
0.194 |
0.018 |
| 53 |
249425_at |
AT5G39790
|
5'-AMP-activated protein kinase-related |
0.193 |
-0.379 |
| 54 |
247024_at |
AT5G66985
|
unknown |
0.192 |
-0.095 |
| 55 |
255782_at |
AT1G19850
|
ARF5, AUXIN RESPONSE FACTOR 5, IAA24, indole-3-acetic acid inducible 24, MP, MONOPTEROS |
0.192 |
0.147 |
| 56 |
245758_at |
AT1G32240
|
KAN2, KANADI 2 |
0.187 |
-0.159 |
| 57 |
255842_at |
AT2G33530
|
scpl46, serine carboxypeptidase-like 46 |
0.187 |
-0.240 |
| 58 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.185 |
0.126 |
| 59 |
261336_at |
AT1G44790
|
[ChaC-like family protein] |
0.185 |
0.015 |
| 60 |
259719_at |
AT1G61070
|
LCR66, low-molecular-weight cysteine-rich 66, PDF2.4, PLANT DEFENSIN 2.4 |
-1.333 |
0.013 |
| 61 |
260038_at |
AT1G68875
|
unknown |
-1.330 |
0.030 |
| 62 |
252063_at |
AT3G51590
|
LTP12, lipid transfer protein 12 |
-1.329 |
0.095 |
| 63 |
250610_at |
AT5G07550
|
ATGRP19, GRP19, glycine-rich protein 19 |
-1.242 |
-0.279 |
| 64 |
260001_at |
AT1G67990
|
ATTSM1, TAPETUM-SPECIFIC METHYLTRANSFERASE 1, TSM1 |
-1.229 |
0.088 |
| 65 |
248350_at |
AT5G52160
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.205 |
0.126 |
| 66 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-1.151 |
0.403 |
| 67 |
262764_at |
AT1G13140
|
CYP86C3, cytochrome P450, family 86, subfamily C, polypeptide 3 |
-1.090 |
0.118 |
| 68 |
260621_at |
AT1G08065
|
ACA5, alpha carbonic anhydrase 5, ATACA5, ALPHA CARBONIC ANHYDRASE 5 |
-1.089 |
0.015 |
| 69 |
256011_at |
AT1G19230
|
[Riboflavin synthase-like superfamily protein] |
-1.032 |
-0.029 |
| 70 |
252035_at |
AT3G52160
|
KCS15, 3-ketoacyl-CoA synthase 15 |
-1.021 |
0.007 |
| 71 |
250576_at |
AT5G08250
|
[Cytochrome P450 superfamily protein] |
-1.020 |
0.155 |
| 72 |
257243_at |
AT3G24230
|
[Pectate lyase family protein] |
-1.013 |
0.043 |
| 73 |
248368_at |
AT5G51950
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.989 |
0.031 |
| 74 |
266530_at |
AT2G16910
|
AMS, ABORTED MICROSPORES |
-0.977 |
0.106 |
| 75 |
248716_at |
AT5G48210
|
unknown |
-0.977 |
0.080 |
| 76 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
-0.903 |
-0.242 |
| 77 |
247727_at |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.898 |
0.132 |
| 78 |
257841_at |
AT3G25260
|
AtNPF4.1, NPF4.1, NRT1/ PTR family 4.1 |
-0.889 |
0.137 |
| 79 |
249881_at |
AT5G23190
|
CYP86B1, cytochrome P450, family 86, subfamily B, polypeptide 1 |
-0.860 |
0.001 |
| 80 |
260791_at |
AT1G06250
|
[alpha/beta-Hydrolases superfamily protein] |
-0.833 |
0.029 |
| 81 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
-0.832 |
-0.189 |
| 82 |
260005_at |
AT1G67920
|
unknown |
-0.823 |
0.164 |
| 83 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-0.814 |
0.132 |
| 84 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
-0.791 |
0.153 |
| 85 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
-0.781 |
0.070 |
| 86 |
256398_at |
AT3G06100
|
NIP7;1, NOD26-like intrinsic protein 7;1, NLM6, NOD26-LIKE MIP 6, NLM8, NOD26-LIKE MIP 8 |
-0.768 |
0.116 |
| 87 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-0.763 |
0.129 |
| 88 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.754 |
-0.048 |
| 89 |
262750_at |
AT1G28710
|
[Nucleotide-diphospho-sugar transferase family protein] |
-0.747 |
0.008 |
| 90 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.740 |
0.158 |
| 91 |
258980_at |
AT3G08900
|
RGP, RGP3, reversibly glycosylated polypeptide 3 |
-0.738 |
0.094 |
| 92 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
-0.735 |
0.140 |
| 93 |
261712_at |
AT1G32780
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.726 |
-0.002 |
| 94 |
253463_at |
AT4G32105
|
[Beta-1,3-N-Acetylglucosaminyltransferase family protein] |
-0.714 |
0.224 |
| 95 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
-0.696 |
0.202 |
| 96 |
246703_at |
AT5G28080
|
WNK9 |
-0.682 |
0.062 |
| 97 |
263757_at |
AT2G21430
|
[Papain family cysteine protease] |
-0.663 |
0.267 |
| 98 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
-0.617 |
-0.131 |
| 99 |
250025_at |
AT5G18290
|
SIP1;2, SIP1B, SMALL AND BASIC INTRINSIC PROTEIN 1B |
-0.615 |
0.145 |
| 100 |
248051_at |
AT5G56110
|
AtMYB103, myb domain protein 103, ATMYB80, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 80, MS188, MALE STERILE 188, MYB103, myb domain protein 103, MYB80 |
-0.608 |
0.051 |
| 101 |
252462_at |
AT3G47250
|
unknown |
-0.604 |
0.075 |
| 102 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
-0.602 |
0.117 |
| 103 |
261567_at |
AT1G33055
|
unknown |
-0.600 |
-0.069 |
| 104 |
261673_at |
AT1G18280
|
LTPG3, glycosylphosphatidylinositol-anchored lipid protein transfer 3 |
-0.588 |
0.157 |
| 105 |
247304_at |
AT5G63850
|
AAP4, amino acid permease 4 |
-0.564 |
0.010 |
| 106 |
248638_at |
AT5G49070
|
KCS21, 3-ketoacyl-CoA synthase 21 |
-0.558 |
0.072 |
| 107 |
250604_at |
AT5G07830
|
AtGUS2, glucuronidase 2, GUS2, glucuronidase 2 |
-0.541 |
-0.140 |
| 108 |
254562_at |
AT4G19230
|
CYP707A1, cytochrome P450, family 707, subfamily A, polypeptide 1 |
-0.541 |
0.258 |
| 109 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
-0.533 |
0.233 |
| 110 |
261392_at |
AT1G79780
|
[Uncharacterised protein family (UPF0497)] |
-0.531 |
0.035 |
| 111 |
259527_at |
AT1G12600
|
[UDP-N-acetylglucosamine (UAA) transporter family] |
-0.520 |
0.108 |
| 112 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
-0.511 |
0.041 |
| 113 |
261396_at |
AT1G79800
|
AtENODL7, ENODL7, early nodulin-like protein 7 |
-0.500 |
0.131 |
| 114 |
260233_at |
AT1G74550
|
CYP98A9, cytochrome P450, family 98, subfamily A, polypeptide 9 |
-0.495 |
0.119 |
| 115 |
248534_at |
AT5G50030
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.491 |
0.071 |
| 116 |
264277_at |
AT1G60390
|
PG1, polygalacturonase 1 |
-0.490 |
0.088 |
| 117 |
260744_at |
AT1G15010
|
unknown |
-0.483 |
0.154 |