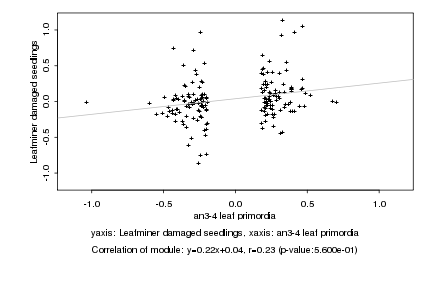
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
an3-4 leaf primordia |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
250135_at |
AT5G15360
|
unknown |
0.697 |
-0.011 |
| 2 |
254566_at |
AT4G19240
|
unknown |
0.673 |
0.009 |
| 3 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.519 |
0.090 |
| 4 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
0.484 |
0.114 |
| 5 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
0.477 |
-0.068 |
| 6 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.463 |
0.186 |
| 7 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.463 |
1.055 |
| 8 |
249970_at |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
0.463 |
0.310 |
| 9 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.459 |
0.180 |
| 10 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
0.443 |
-0.056 |
| 11 |
253532_at |
AT4G31570
|
unknown |
0.411 |
-0.138 |
| 12 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.405 |
0.971 |
| 13 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.392 |
-0.126 |
| 14 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.390 |
0.174 |
| 15 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.387 |
0.150 |
| 16 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.387 |
0.205 |
| 17 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.386 |
0.182 |
| 18 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.386 |
0.172 |
| 19 |
262421_at |
AT1G50290
|
unknown |
0.383 |
-0.009 |
| 20 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
0.382 |
-0.132 |
| 21 |
248810_at |
AT5G47280
|
ADR1-L3, ADR1-like 3 |
0.367 |
-0.036 |
| 22 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.354 |
0.549 |
| 23 |
253172_at |
AT4G35060
|
HIPP25, heavy metal associated isoprenylated plant protein 25 |
0.353 |
0.444 |
| 24 |
257952_at |
AT3G21770
|
[Peroxidase superfamily protein] |
0.347 |
-0.040 |
| 25 |
265044_at |
AT1G03950
|
VPS2.3, vacuolar protein sorting-associated protein 2.3 |
0.341 |
0.135 |
| 26 |
256976_at |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
0.335 |
-0.079 |
| 27 |
265025_at |
AT1G24575
|
unknown |
0.328 |
0.240 |
| 28 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
0.327 |
1.146 |
| 29 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
0.327 |
-0.432 |
| 30 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.315 |
0.928 |
| 31 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
0.313 |
-0.112 |
| 32 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
0.307 |
-0.446 |
| 33 |
260991_at |
AT1G12160
|
[Flavin-binding monooxygenase family protein] |
0.305 |
0.056 |
| 34 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.304 |
0.139 |
| 35 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.303 |
0.395 |
| 36 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
0.298 |
0.071 |
| 37 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
0.282 |
0.020 |
| 38 |
246868_at |
AT5G26010
|
[Protein phosphatase 2C family protein] |
0.280 |
0.083 |
| 39 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.272 |
0.156 |
| 40 |
261745_at |
AT1G08500
|
AtENODL18, ENODL18, early nodulin-like protein 18 |
0.272 |
0.118 |
| 41 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.266 |
-0.175 |
| 42 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.264 |
-0.339 |
| 43 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
0.262 |
0.089 |
| 44 |
261845_at |
AT1G15960
|
ATNRAMP6, NRAMP6, NRAMP metal ion transporter 6 |
0.260 |
-0.211 |
| 45 |
252765_at |
AT3G42800
|
unknown |
0.257 |
-0.176 |
| 46 |
253689_at |
AT4G29770
|
unknown |
0.256 |
-0.047 |
| 47 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
0.255 |
0.416 |
| 48 |
252495_at |
AT3G46770
|
[AP2/B3-like transcriptional factor family protein] |
0.252 |
-0.103 |
| 49 |
262236_at |
AT1G48330
|
unknown |
0.247 |
0.278 |
| 50 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.243 |
-0.096 |
| 51 |
257493_at |
AT1G48180
|
unknown |
0.243 |
0.010 |
| 52 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
0.242 |
-0.044 |
| 53 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
0.239 |
0.011 |
| 54 |
256120_at |
AT1G18130
|
[Class II aaRS and biotin synthetases superfamily protein] |
0.238 |
0.009 |
| 55 |
260396_at |
AT1G69720
|
HO3, heme oxygenase 3 |
0.238 |
0.125 |
| 56 |
244921_s_at |
ATMG01000
|
ORF114 |
0.232 |
0.135 |
| 57 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
0.232 |
0.566 |
| 58 |
244902_at |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
0.231 |
0.042 |
| 59 |
257334_at |
ATMG01370
|
ORF111D |
0.231 |
0.079 |
| 60 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
0.224 |
0.037 |
| 61 |
254596_at |
AT4G18975
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.223 |
-0.016 |
| 62 |
266613_at |
AT2G14900
|
[Gibberellin-regulated family protein] |
0.222 |
-0.180 |
| 63 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.218 |
0.413 |
| 64 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
0.218 |
-0.054 |
| 65 |
262503_at |
AT1G21670
|
[LOCATED IN: cell wall, plant-type cell wall] |
0.217 |
0.247 |
| 66 |
252419_at |
AT3G47510
|
unknown |
0.214 |
0.198 |
| 67 |
246168_at |
AT5G32460
|
[Transcriptional factor B3 family protein] |
0.213 |
-0.016 |
| 68 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
0.213 |
-0.050 |
| 69 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
0.212 |
0.011 |
| 70 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
0.211 |
0.041 |
| 71 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
0.206 |
-0.277 |
| 72 |
260412_at |
AT1G69830
|
AMY3, alpha-amylase-like 3, ATAMY3, ALPHA-AMYLASE-LIKE 3 |
0.206 |
0.293 |
| 73 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
0.205 |
-0.095 |
| 74 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
0.205 |
0.053 |
| 75 |
251372_at |
AT3G60520
|
unknown |
0.204 |
0.244 |
| 76 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
0.203 |
-0.190 |
| 77 |
256145_at |
AT1G48750
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.202 |
-0.077 |
| 78 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
0.201 |
-0.137 |
| 79 |
250872_at |
AT5G03960
|
IQD12, IQ-domain 12 |
0.200 |
0.049 |
| 80 |
263688_at |
AT1G26920
|
unknown |
0.198 |
0.164 |
| 81 |
260352_at |
AT1G69295
|
PDCB4, plasmodesmata callose-binding protein 4 |
0.197 |
-0.002 |
| 82 |
255980_at |
AT1G33970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.194 |
0.462 |
| 83 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
0.192 |
0.382 |
| 84 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.190 |
0.250 |
| 85 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
0.186 |
0.647 |
| 86 |
259680_at |
AT1G77690
|
LAX3, like AUX1 3 |
0.186 |
-0.370 |
| 87 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
0.186 |
0.461 |
| 88 |
245197_at |
AT1G67800
|
[Copine (Calcium-dependent phospholipid-binding protein) family] |
0.184 |
0.136 |
| 89 |
257158_at |
AT3G24360
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.179 |
-0.121 |
| 90 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.178 |
0.184 |
| 91 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.178 |
0.402 |
| 92 |
255726_at |
AT1G25530
|
[Transmembrane amino acid transporter family protein] |
0.175 |
-0.298 |
| 93 |
246103_at |
AT5G28640
|
AN3, ANGUSTIFOLIA 3, ATGIF1, ARABIDOPSIS GRF1-INTERACTING FACTOR 1, GIF, GRF1-INTERACTING FACTOR, GIF1, GRF1-INTERACTING FACTOR 1 |
-1.039 |
-0.006 |
| 94 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.600 |
-0.027 |
| 95 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.552 |
-0.181 |
| 96 |
258424_at |
AT3G16750
|
unknown |
-0.508 |
-0.162 |
| 97 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
-0.498 |
0.023 |
| 98 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.477 |
-0.204 |
| 99 |
252659_at |
AT3G44430
|
unknown |
-0.467 |
-0.076 |
| 100 |
262458_at |
AT1G11280
|
[S-locus lectin protein kinase family protein] |
-0.461 |
-0.129 |
| 101 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.443 |
-0.116 |
| 102 |
255484_at |
AT4G02540
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.443 |
-0.155 |
| 103 |
263023_at |
AT1G23960
|
unknown |
-0.437 |
0.040 |
| 104 |
257579_at |
AT3G11000
|
[DCD (Development and Cell Death) domain protein] |
-0.435 |
0.020 |
| 105 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.433 |
0.749 |
| 106 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.421 |
0.086 |
| 107 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.419 |
-0.277 |
| 108 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
-0.418 |
-0.168 |
| 109 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
-0.412 |
-0.099 |
| 110 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
-0.411 |
0.053 |
| 111 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
-0.406 |
-0.100 |
| 112 |
256568_at |
AT3G19520
|
unknown |
-0.399 |
0.041 |
| 113 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-0.393 |
-0.143 |
| 114 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.375 |
0.073 |
| 115 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.369 |
-0.277 |
| 116 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
-0.363 |
0.514 |
| 117 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.362 |
-0.319 |
| 118 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
-0.361 |
0.020 |
| 119 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.361 |
0.224 |
| 120 |
256924_at |
AT3G29590
|
AT5MAT |
-0.355 |
0.013 |
| 121 |
260975_at |
AT1G53430
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.349 |
0.212 |
| 122 |
246393_at |
AT1G58150
|
unknown |
-0.347 |
-0.200 |
| 123 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.345 |
-0.358 |
| 124 |
259637_at |
AT1G52260
|
ATPDI3, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 3, ATPDIL1-5, PDI-like 1-5, PDI3, PROTEIN DISULFIDE ISOMERASE 3, PDIL1-5, PDI-like 1-5 |
-0.341 |
-0.069 |
| 125 |
252593_at |
AT3G45590
|
ATSEN1, splicing endonuclease 1, SEN1, splicing endonuclease 1 |
-0.330 |
0.107 |
| 126 |
262575_at |
AT1G15210
|
ABCG35, ATP-binding cassette G35, ATPDR7, PLEIOTROPIC DRUG RESISTANCE 7, PDR7, pleiotropic drug resistance 7 |
-0.329 |
0.076 |
| 127 |
256096_at |
AT1G13650
|
unknown |
-0.328 |
-0.608 |
| 128 |
264156_at |
AT1G65280
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.328 |
-0.028 |
| 129 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.325 |
0.065 |
| 130 |
255044_at |
AT4G09680
|
ATCTC1, CTC1, conserved telomere maintenance component 1 |
-0.322 |
-0.079 |
| 131 |
258159_at |
AT3G17840
|
RLK902, receptor-like kinase 902 |
-0.312 |
-0.505 |
| 132 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
-0.307 |
-0.010 |
| 133 |
259549_at |
AT1G35290
|
ALT1, acyl-lipid thioesterase 1 |
-0.301 |
-0.032 |
| 134 |
252652_at |
AT3G44720
|
ADT4, arogenate dehydratase 4 |
-0.299 |
0.272 |
| 135 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
-0.296 |
0.727 |
| 136 |
259110_at |
AT3G05570
|
unknown |
-0.294 |
0.107 |
| 137 |
256303_at |
AT1G69550
|
[disease resistance protein (TIR-NBS-LRR class)] |
-0.292 |
-0.016 |
| 138 |
255260_at |
AT4G05040
|
[ankyrin repeat family protein] |
-0.292 |
-0.236 |
| 139 |
252432_at |
AT3G47680
|
[DNA binding] |
-0.286 |
-0.023 |
| 140 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.282 |
0.437 |
| 141 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
-0.280 |
0.020 |
| 142 |
262346_at |
AT1G63980
|
[D111/G-patch domain-containing protein] |
-0.279 |
0.025 |
| 143 |
256875_at |
AT3G26330
|
CYP71B37, cytochrome P450, family 71, subfamily B, polypeptide 37 |
-0.275 |
0.381 |
| 144 |
259537_at |
AT1G12370
|
PHR1, photolyase 1, UVR2, UV RESISTANCE 2 |
-0.265 |
-0.252 |
| 145 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
-0.265 |
0.029 |
| 146 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.262 |
-0.113 |
| 147 |
247592_at |
AT5G60780
|
ATNRT2.3, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 2.3, NRT2.3, nitrate transporter 2.3 |
-0.262 |
-0.016 |
| 148 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.260 |
-0.864 |
| 149 |
250537_at |
AT5G08565
|
[Transcription initiation Spt4-like protein] |
-0.252 |
0.200 |
| 150 |
262028_at |
AT1G35560
|
AtTCP23, TCP23, TCP domain protein 23 |
-0.252 |
-0.132 |
| 151 |
262649_at |
AT1G14040
|
[EXS (ERD1/XPR1/SYG1) family protein] |
-0.249 |
0.029 |
| 152 |
258027_at |
AT3G19515
|
unknown |
-0.247 |
-0.197 |
| 153 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.244 |
-0.745 |
| 154 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.244 |
0.967 |
| 155 |
264658_at |
AT1G09910
|
[Rhamnogalacturonate lyase family protein] |
-0.243 |
0.043 |
| 156 |
259686_at |
AT1G63100
|
[GRAS family transcription factor] |
-0.243 |
-0.010 |
| 157 |
247317_at |
AT5G64060
|
anac103, NAC domain containing protein 103, NAC103, NAC domain containing protein 103 |
-0.241 |
-0.062 |
| 158 |
254585_at |
AT4G19500
|
[nucleoside-triphosphatases] |
-0.240 |
-0.215 |
| 159 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
-0.238 |
-0.218 |
| 160 |
246320_at |
AT1G16560
|
[Per1-like family protein] |
-0.238 |
0.092 |
| 161 |
257013_at |
AT3G26922
|
[F-box/RNI-like superfamily protein] |
-0.237 |
-0.048 |
| 162 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
-0.237 |
0.291 |
| 163 |
248335_at |
AT5G52450
|
[MATE efflux family protein] |
-0.236 |
0.099 |
| 164 |
261642_at |
AT1G27680
|
APL2, ADPGLC-PPase large subunit |
-0.234 |
-0.051 |
| 165 |
265441_at |
AT2G20870
|
[cell wall protein precursor, putative] |
-0.234 |
0.274 |
| 166 |
252669_at |
AT3G44100
|
[MD-2-related lipid recognition domain-containing protein] |
-0.233 |
0.060 |
| 167 |
257554_at |
AT3G24890
|
ATVAMP728, VAMP728, vesicle-associated membrane protein 728 |
-0.229 |
0.002 |
| 168 |
264987_at |
AT1G27030
|
unknown |
-0.227 |
-0.070 |
| 169 |
262908_at |
AT1G59900
|
AT-E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit, E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit |
-0.226 |
0.004 |
| 170 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.220 |
-0.402 |
| 171 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.218 |
0.538 |
| 172 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.217 |
0.105 |
| 173 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.215 |
-0.461 |
| 174 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
-0.211 |
-0.043 |
| 175 |
250755_at |
AT5G05750
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.208 |
0.065 |
| 176 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-0.208 |
-0.383 |
| 177 |
252646_at |
AT3G44610
|
[Protein kinase superfamily protein] |
-0.207 |
-0.111 |
| 178 |
260502_at |
AT1G47270
|
AtTLP6, tubby like protein 6, TLP6, tubby like protein 6 |
-0.205 |
-0.125 |
| 179 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.205 |
-0.730 |
| 180 |
245313_at |
AT4G15420
|
[Ubiquitin fusion degradation UFD1 family protein] |
-0.204 |
0.043 |
| 181 |
258326_at |
AT3G22760
|
SOL1 |
-0.202 |
-0.312 |
| 182 |
259574_at |
AT1G35310
|
MLP168, MLP-like protein 168 |
-0.201 |
-0.004 |
| 183 |
259934_at |
AT1G71340
|
AtGDPD4, GDPD4, glycerophosphodiester phosphodiesterase 4 |
-0.200 |
-0.306 |