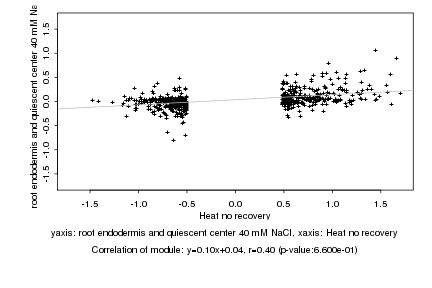
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Heat no recovery |
Log2 signal ratio
root endodermis and quiescent center 40 mM NaCl |
| 1 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
1.699 |
0.180 |
| 2 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.654 |
0.897 |
| 3 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
1.603 |
-0.056 |
| 4 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.592 |
0.580 |
| 5 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
1.564 |
0.199 |
| 6 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
1.553 |
0.350 |
| 7 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
1.480 |
0.115 |
| 8 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
1.448 |
0.042 |
| 9 |
244931_at |
ATMG00630
|
ORF110B |
1.441 |
0.034 |
| 10 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.438 |
1.066 |
| 11 |
257323_at |
ATMG01200
|
ORF294 |
1.432 |
0.152 |
| 12 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
1.396 |
0.254 |
| 13 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
1.382 |
0.337 |
| 14 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
1.362 |
0.269 |
| 15 |
244902_at |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
1.332 |
0.046 |
| 16 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
1.326 |
0.644 |
| 17 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
1.307 |
0.236 |
| 18 |
257337_at |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
1.300 |
0.045 |
| 19 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
1.298 |
0.411 |
| 20 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
1.295 |
0.180 |
| 21 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
1.287 |
0.626 |
| 22 |
244901_at |
ATMG00640
|
ORF25 |
1.283 |
0.071 |
| 23 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
1.276 |
0.156 |
| 24 |
255811_at |
AT4G10250
|
ATHSP22.0 |
1.253 |
0.200 |
| 25 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
1.208 |
0.028 |
| 26 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
1.200 |
0.141 |
| 27 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
1.197 |
-0.046 |
| 28 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
1.197 |
0.004 |
| 29 |
258695_at |
AT3G09640
|
APX1B, ASCORBATE PEROXIDASE 1B, APX2, ascorbate peroxidase 2 |
1.171 |
0.037 |
| 30 |
263062_at |
AT2G18180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
1.169 |
-0.002 |
| 31 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
1.144 |
0.248 |
| 32 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
1.144 |
0.579 |
| 33 |
248752_at |
AT5G47600
|
[HSP20-like chaperones superfamily protein] |
1.139 |
-0.084 |
| 34 |
263061_at |
AT2G18190
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
1.138 |
0.191 |
| 35 |
250826_at |
AT5G05220
|
unknown |
1.136 |
0.239 |
| 36 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.133 |
0.496 |
| 37 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
1.130 |
-0.036 |
| 38 |
263881_at |
AT2G21820
|
unknown |
1.127 |
0.377 |
| 39 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
1.115 |
0.234 |
| 40 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
1.100 |
0.241 |
| 41 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
1.095 |
0.267 |
| 42 |
257321_at |
ATMG01130
|
ORF106F |
1.083 |
0.029 |
| 43 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
1.080 |
0.309 |
| 44 |
255246_at |
AT4G05640
|
unknown |
1.078 |
0.058 |
| 45 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.068 |
0.127 |
| 46 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
1.062 |
0.262 |
| 47 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
1.061 |
0.027 |
| 48 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
1.054 |
0.494 |
| 49 |
245703_at |
AT5G04380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
1.053 |
0.021 |
| 50 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
1.044 |
0.010 |
| 51 |
257322_at |
ATMG01180
|
ORF111B |
1.037 |
0.023 |
| 52 |
256603_at |
AT3G28270
|
unknown |
1.037 |
-0.001 |
| 53 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
1.034 |
0.602 |
| 54 |
247447_at |
AT5G62730
|
[Major facilitator superfamily protein] |
1.029 |
0.034 |
| 55 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
1.026 |
0.050 |
| 56 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
1.025 |
0.054 |
| 57 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
1.018 |
0.156 |
| 58 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
1.011 |
0.175 |
| 59 |
257317_at |
ATMG01060
|
ORF107G |
1.008 |
0.011 |
| 60 |
265024_at |
AT1G24600
|
unknown |
0.993 |
0.369 |
| 61 |
250624_at |
AT5G07330
|
unknown |
0.975 |
0.471 |
| 62 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.973 |
0.074 |
| 63 |
248218_at |
AT5G53710
|
unknown |
0.966 |
0.171 |
| 64 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.960 |
0.805 |
| 65 |
244923_s_at |
ATMG01020
|
ORF153B |
0.952 |
0.064 |
| 66 |
249614_at |
AT5G37300
|
WSD1 |
0.952 |
0.054 |
| 67 |
244904_at |
ATMG00670
|
ORF275 |
0.947 |
0.055 |
| 68 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.945 |
0.207 |
| 69 |
244906_at |
ATMG00690
|
ORF240A |
0.936 |
-0.041 |
| 70 |
256114_at |
AT1G16850
|
unknown |
0.934 |
0.588 |
| 71 |
244903_at |
ATMG00660
|
ORF149 |
0.932 |
0.142 |
| 72 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.929 |
0.278 |
| 73 |
257332_at |
ATMG01350
|
ORF145C |
0.922 |
0.043 |
| 74 |
246125_at |
AT5G19875
|
unknown |
0.921 |
0.310 |
| 75 |
264580_at |
AT1G05340
|
unknown |
0.914 |
0.316 |
| 76 |
259054_at |
AT3G03480
|
CHAT, acetyl CoA:(Z)-3-hexen-1-ol acetyltransferase |
0.911 |
0.018 |
| 77 |
265242_at |
AT2G07705
|
unknown |
0.908 |
0.051 |
| 78 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.902 |
0.174 |
| 79 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
0.899 |
-0.192 |
| 80 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.894 |
0.519 |
| 81 |
244921_s_at |
ATMG01000
|
ORF114 |
0.891 |
0.080 |
| 82 |
258397_at |
AT3G15357
|
unknown |
0.891 |
-0.048 |
| 83 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.890 |
0.596 |
| 84 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.874 |
0.162 |
| 85 |
258325_at |
AT3G22830
|
AT-HSFA6B, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A6B, HSFA6B, heat shock transcription factor A6B |
0.869 |
0.072 |
| 86 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
0.867 |
0.051 |
| 87 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
0.866 |
0.097 |
| 88 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.864 |
0.031 |
| 89 |
250162_at |
AT5G15250
|
ATFTSH6, FTSH6, FTSH protease 6 |
0.856 |
0.129 |
| 90 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.855 |
0.268 |
| 91 |
254263_at |
AT4G23493
|
unknown |
0.848 |
0.032 |
| 92 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.846 |
0.182 |
| 93 |
254413_at |
AT4G21440
|
ATM4, A. THALIANA MYB 4, ATMYB102, MYB-like 102, MYB102, MYB102, MYB-like 102 |
0.845 |
0.151 |
| 94 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.837 |
0.084 |
| 95 |
257336_at |
ATMG01410
|
ORF204, open reading frame 204 |
0.832 |
0.031 |
| 96 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.830 |
0.248 |
| 97 |
265483_at |
AT2G15790
|
CYP40, CYCLOPHILIN 40, SQN, SQUINT |
0.830 |
0.110 |
| 98 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
0.830 |
0.011 |
| 99 |
254574_at |
AT4G19430
|
unknown |
0.826 |
0.026 |
| 100 |
266959_at |
AT2G07690
|
MCM5, MINICHROMOSOME MAINTENANCE 5 |
0.826 |
-0.043 |
| 101 |
245903_at |
AT5G11100
|
ATSYTD, NTMC2T2.2, NTMC2TYPE2.2, SYT4, synaptotagmin 4, SYTD |
0.821 |
-0.001 |
| 102 |
255048_at |
AT4G09600
|
GASA3, GAST1 protein homolog 3 |
0.818 |
0.058 |
| 103 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.817 |
0.058 |
| 104 |
244925_at |
ATMG00510
|
NAD7, NADH dehydrogenase subunit 7 |
0.813 |
0.047 |
| 105 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.808 |
0.217 |
| 106 |
244945_at |
ATMG00110
|
ABCI2, ATP-binding cassette I2, CCB206, cytochrome C biogenesis 206 |
0.801 |
-0.010 |
| 107 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.798 |
0.545 |
| 108 |
244958_at |
ATMG00450
|
ORF106B |
0.798 |
0.086 |
| 109 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.791 |
-0.179 |
| 110 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
0.788 |
0.069 |
| 111 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.788 |
0.254 |
| 112 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.785 |
-0.069 |
| 113 |
249913_at |
AT5G22810
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.778 |
0.048 |
| 114 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.776 |
0.459 |
| 115 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.776 |
0.419 |
| 116 |
246242_at |
AT4G36600
|
[Late embryogenesis abundant (LEA) protein] |
0.772 |
0.084 |
| 117 |
248959_at |
AT5G45630
|
unknown |
0.765 |
0.005 |
| 118 |
249837_at |
AT5G23480
|
[SWIB/MDM2 domain] |
0.763 |
0.011 |
| 119 |
257035_at |
AT3G19270
|
CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4 |
0.757 |
0.027 |
| 120 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
0.757 |
0.290 |
| 121 |
256779_at |
AT3G13784
|
AtcwINV5, cell wall invertase 5, CWINV5, cell wall invertase 5 |
0.756 |
0.027 |
| 122 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
0.756 |
0.137 |
| 123 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.754 |
-0.093 |
| 124 |
260917_at |
AT1G02700
|
unknown |
0.742 |
0.043 |
| 125 |
265234_at |
AT2G07721
|
unknown |
0.742 |
0.052 |
| 126 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.740 |
0.091 |
| 127 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.738 |
0.104 |
| 128 |
253019_at |
AT4G38010
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.736 |
0.078 |
| 129 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.730 |
0.023 |
| 130 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.721 |
0.143 |
| 131 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.711 |
0.136 |
| 132 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.710 |
0.314 |
| 133 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
0.710 |
0.282 |
| 134 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.706 |
-0.066 |
| 135 |
256518_at |
AT1G66080
|
unknown |
0.706 |
0.084 |
| 136 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.704 |
0.061 |
| 137 |
254350_at |
AT4G22280
|
[F-box/RNI-like superfamily protein] |
0.698 |
-0.032 |
| 138 |
263515_at |
AT2G21640
|
unknown |
0.688 |
0.074 |
| 139 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
0.687 |
-0.002 |
| 140 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
0.684 |
0.102 |
| 141 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.682 |
-0.032 |
| 142 |
262459_at |
AT1G50400
|
[Eukaryotic porin family protein] |
0.680 |
-0.037 |
| 143 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.677 |
0.095 |
| 144 |
257334_at |
ATMG01370
|
ORF111D |
0.673 |
-0.009 |
| 145 |
257874_at |
AT3G17110
|
[pseudogene] |
0.669 |
0.177 |
| 146 |
249439_at |
AT5G40020
|
[Pathogenesis-related thaumatin superfamily protein] |
0.665 |
-0.305 |
| 147 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
0.661 |
0.237 |
| 148 |
258406_at |
AT3G17611
|
ATRBL10, RHOMBOID-like protein 10, ATRBL14, RHOMBOID-like protein 14, RBL10, RHOMBOID-like protein 10, RBL14, RHOMBOID-like protein 14 |
0.660 |
0.097 |
| 149 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.652 |
0.136 |
| 150 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
0.651 |
0.306 |
| 151 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.651 |
-0.201 |
| 152 |
250103_at |
AT5G16600
|
AtMYB43, myb domain protein 43, MYB43, myb domain protein 43 |
0.649 |
0.083 |
| 153 |
251896_at |
AT3G54390
|
[sequence-specific DNA binding transcription factors] |
0.649 |
0.029 |
| 154 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
0.648 |
-0.083 |
| 155 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
0.644 |
0.038 |
| 156 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.644 |
0.096 |
| 157 |
256248_at |
AT3G66652
|
[fip1 motif-containing protein] |
0.643 |
0.077 |
| 158 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
0.639 |
0.019 |
| 159 |
244941_at |
ATMG00010
|
ORF153A |
0.637 |
-0.020 |
| 160 |
266046_at |
AT2G07728
|
unknown |
0.637 |
-0.010 |
| 161 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.634 |
0.000 |
| 162 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.634 |
-0.032 |
| 163 |
257853_at |
AT3G12960
|
SMP1, seed maturation protein 1 |
0.629 |
0.017 |
| 164 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
0.626 |
0.206 |
| 165 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.623 |
0.564 |
| 166 |
250660_at |
AT5G07060
|
MAC5C, MOS4-associated complex subunit 5C |
0.623 |
-0.008 |
| 167 |
256852_at |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
0.622 |
0.315 |
| 168 |
250722_at |
AT5G06190
|
unknown |
0.622 |
0.101 |
| 169 |
257207_at |
AT3G14900
|
EMB3120, EMBRYO DEFECTIVE 3120 |
0.621 |
0.024 |
| 170 |
250910_at |
AT5G03720
|
AT-HSFA3, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A3, HSFA3, heat shock transcription factor A3 |
0.615 |
0.286 |
| 171 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
0.613 |
0.410 |
| 172 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.610 |
0.219 |
| 173 |
252770_at |
AT3G42860
|
[zinc knuckle (CCHC-type) family protein] |
0.609 |
0.227 |
| 174 |
253432_at |
AT4G32450
|
MEF8S, MEF8 similar |
0.608 |
0.126 |
| 175 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
0.607 |
0.132 |
| 176 |
249598_at |
AT5G37970
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.607 |
-0.040 |
| 177 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
0.599 |
-0.060 |
| 178 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
0.599 |
0.230 |
| 179 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
0.599 |
0.035 |
| 180 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.594 |
0.185 |
| 181 |
251985_at |
AT3G53220
|
[Thioredoxin superfamily protein] |
0.592 |
0.014 |
| 182 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
0.591 |
-0.119 |
| 183 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
0.587 |
0.260 |
| 184 |
265276_at |
AT2G28400
|
unknown |
0.587 |
0.116 |
| 185 |
247119_at |
AT5G65900
|
[DEA(D/H)-box RNA helicase family protein] |
0.582 |
0.007 |
| 186 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.582 |
0.328 |
| 187 |
254639_at |
AT4G18750
|
DOT4, DEFECTIVELY ORGANIZED TRIBUTARIES 4 |
0.578 |
0.060 |
| 188 |
264948_at |
AT1G77030
|
[hydrolases, acting on acid anhydrides, in phosphorus-containing anhydrides] |
0.577 |
-0.001 |
| 189 |
244918_at |
ATMG00890
|
ORF106D |
0.577 |
0.022 |
| 190 |
256416_at |
AT3G11050
|
ATFER2, ferritin 2, FER2, ferritin 2 |
0.576 |
0.056 |
| 191 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
0.575 |
0.060 |
| 192 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.575 |
0.137 |
| 193 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.573 |
0.012 |
| 194 |
250087_at |
AT5G17270
|
[Protein prenylyltransferase superfamily protein] |
0.572 |
0.019 |
| 195 |
264972_at |
AT1G67370
|
ASY1, ASYNAPTIC 1, ATASY1 |
0.571 |
0.181 |
| 196 |
250036_at |
AT5G18340
|
[ARM repeat superfamily protein] |
0.571 |
0.095 |
| 197 |
247659_at |
AT5G60040
|
NRPC1, nuclear RNA polymerase C1 |
0.571 |
0.003 |
| 198 |
254629_at |
AT4G18425
|
unknown |
0.571 |
-0.130 |
| 199 |
255494_at |
AT4G02710
|
NET1C, Networked 1C |
0.570 |
-0.056 |
| 200 |
261655_at |
AT1G01940
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.568 |
0.042 |
| 201 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.564 |
0.080 |
| 202 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.564 |
0.209 |
| 203 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.563 |
0.252 |
| 204 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.563 |
0.087 |
| 205 |
248505_at |
AT5G50360
|
unknown |
0.561 |
0.317 |
| 206 |
267309_at |
AT2G19385
|
[zinc ion binding] |
0.561 |
-0.059 |
| 207 |
246196_at |
AT4G37090
|
unknown |
0.560 |
0.009 |
| 208 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.558 |
0.088 |
| 209 |
258794_at |
AT3G04710
|
TPR10, tetratricopeptide repeat 10 |
0.558 |
0.050 |
| 210 |
263164_at |
AT1G03070
|
AtLFG4, LFG4, LIFEGUARD 4 |
0.554 |
0.098 |
| 211 |
262641_at |
AT1G62730
|
[Terpenoid synthases superfamily protein] |
0.554 |
0.054 |
| 212 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
0.553 |
0.041 |
| 213 |
251652_at |
AT3G57380
|
[Glycosyltransferase family 61 protein] |
0.552 |
0.118 |
| 214 |
259489_at |
AT1G15790
|
unknown |
0.552 |
0.019 |
| 215 |
265931_at |
AT2G18520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.551 |
0.054 |
| 216 |
249019_at |
AT5G44780
|
MORF4, multiple organellar RNA editing factor 4 |
0.550 |
-0.067 |
| 217 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
0.550 |
-0.143 |
| 218 |
250504_at |
AT5G09840
|
[Putative endonuclease or glycosyl hydrolase] |
0.550 |
-0.054 |
| 219 |
264331_at |
AT1G04130
|
AtTPR2, TPR2, tetratricopeptide repeat 2 |
0.546 |
0.071 |
| 220 |
254390_at |
AT4G21940
|
CPK15, calcium-dependent protein kinase 15 |
0.546 |
-0.041 |
| 221 |
259503_at |
AT1G15870
|
[Mitochondrial glycoprotein family protein] |
0.546 |
-0.028 |
| 222 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.545 |
-0.090 |
| 223 |
247649_at |
AT5G60030
|
unknown |
0.543 |
-0.014 |
| 224 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.543 |
0.238 |
| 225 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
0.542 |
0.127 |
| 226 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.541 |
-0.320 |
| 227 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
0.541 |
0.067 |
| 228 |
250762_at |
AT5G05990
|
[Mitochondrial glycoprotein family protein] |
0.540 |
-0.156 |
| 229 |
244942_at |
ATMG00050
|
ORF131 |
0.539 |
0.082 |
| 230 |
257859_at |
AT3G12955
|
SAUR74, SMALL AUXIN UPREGULATED RNA 74 |
0.537 |
0.068 |
| 231 |
257822_at |
AT3G25230
|
ATFKBP62, FKBP62, FK506 BINDING PROTEIN 62, ROF1, rotamase FKBP 1 |
0.537 |
0.049 |
| 232 |
245879_at |
AT5G09420
|
AtmtOM64, ATTOC64-V, translocon at the outer membrane of chloroplasts 64-V, MTOM64, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-V, OM64, outer membrane 64, TOC64-V, translocon at the outer membrane of chloroplasts 64-V |
0.537 |
0.011 |
| 233 |
245904_at |
AT5G11110
|
ATSPS2F, sucrose phosphate synthase 2F, KNS2, KAONASHI 2, SPS1, SUCROSE PHOSPHATE SYNTHASE 1, SPS2F, sucrose phosphate synthase 2F, SPSA2, sucrose-phosphate synthase A2 |
0.537 |
0.383 |
| 234 |
267110_at |
AT2G14800
|
unknown |
0.537 |
0.097 |
| 235 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
0.535 |
0.059 |
| 236 |
256027_at |
AT1G34160
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.533 |
0.084 |
| 237 |
251049_at |
AT5G02430
|
[Transducin/WD40 repeat-like superfamily protein] |
0.530 |
0.066 |
| 238 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
0.528 |
0.140 |
| 239 |
255481_at |
AT4G02460
|
AtPMS1, PMS1, POSTMEIOTIC SEGREGATION 1 |
0.528 |
-0.025 |
| 240 |
258336_at |
AT3G16050
|
A37, ATPDX1.2, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.2, PDX1.2, pyridoxine biosynthesis 1.2 |
0.528 |
-0.015 |
| 241 |
250326_at |
AT5G12080
|
ATMSL10, MSL10, mechanosensitive channel of small conductance-like 10 |
0.528 |
0.158 |
| 242 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
0.528 |
-0.159 |
| 243 |
254076_at |
AT4G25340
|
ATFKBP53, FKBP53, FK506 BINDING PROTEIN 53 |
0.527 |
0.033 |
| 244 |
252962_at |
AT4G38780
|
[Pre-mRNA-processing-splicing factor] |
0.527 |
-0.103 |
| 245 |
265463_at |
AT2G37090
|
IRX9, IRREGULAR XYLEM 9 |
0.527 |
-0.286 |
| 246 |
264142_at |
AT1G78930
|
[Mitochondrial transcription termination factor family protein] |
0.527 |
-0.030 |
| 247 |
245243_at |
AT1G44414
|
unknown |
0.526 |
0.067 |
| 248 |
246581_at |
AT1G31760
|
[SWIB/MDM2 domain superfamily protein] |
0.524 |
-0.004 |
| 249 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
0.523 |
0.382 |
| 250 |
258506_at |
AT3G06520
|
[agenet domain-containing protein] |
0.522 |
0.080 |
| 251 |
259248_at |
AT3G07770
|
AtHsp90-6, HEAT SHOCK PROTEIN 90-6, AtHsp90.6, HEAT SHOCK PROTEIN 90.6, Hsp89.1, HEAT SHOCK PROTEIN 89.1 |
0.522 |
-0.019 |
| 252 |
264150_at |
AT1G02090
|
ATCSN7, ARABIDOPSIS THALIANA COP9 SIGNALOSOME SUBUNIT 7, COP15, CONSTITUTIVE PHOTOMORPHOGENIC 15, CSN7, COP9 SIGNALOSOME SUBUNIT 7, FUS5, FUSCA 5 |
0.521 |
0.076 |
| 253 |
257694_at |
AT3G12860
|
[NOP56-like pre RNA processing ribonucleoprotein] |
0.521 |
-0.038 |
| 254 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.520 |
0.544 |
| 255 |
247234_at |
AT5G64980
|
unknown |
0.520 |
0.008 |
| 256 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.519 |
0.076 |
| 257 |
247491_at |
AT5G61880
|
[Protein Transporter, Pam16] |
0.519 |
0.055 |
| 258 |
247893_at |
AT5G57980
|
RPB5C, RNA polymerase II fifth largest subunit, C |
0.516 |
-0.165 |
| 259 |
244948_at |
ATMG00140
|
ORF167 |
0.516 |
0.101 |
| 260 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
0.513 |
0.075 |
| 261 |
251678_at |
AT3G56990
|
EDA7, embryo sac development arrest 7 |
0.513 |
-0.016 |
| 262 |
258871_at |
AT3G03060
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.513 |
-0.059 |
| 263 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
0.510 |
-0.004 |
| 264 |
267627_at |
AT2G42270
|
[U5 small nuclear ribonucleoprotein helicase] |
0.510 |
0.199 |
| 265 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.509 |
0.144 |
| 266 |
264814_at |
AT2G17900
|
ASHR1, ASH1-related 1, SDG37, SET domain group 37 |
0.508 |
0.099 |
| 267 |
250211_at |
AT5G13880
|
unknown |
0.506 |
0.390 |
| 268 |
251964_at |
AT3G53370
|
[S1FA-like DNA-binding protein] |
0.505 |
-0.087 |
| 269 |
257765_at |
AT3G23020
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.505 |
0.004 |
| 270 |
253777_at |
AT4G28450
|
[nucleotide binding] |
0.502 |
-0.013 |
| 271 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
0.502 |
-0.099 |
| 272 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.501 |
0.034 |
| 273 |
253638_at |
AT4G30470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.501 |
0.040 |
| 274 |
267081_at |
AT2G41210
|
PIP5K5, phosphatidylinositol- 4-phosphate 5-kinase 5 |
0.501 |
0.148 |
| 275 |
265088_at |
AT1G03750
|
CHR9, CHROMATIN REMODELING 9 |
0.500 |
0.115 |
| 276 |
264968_at |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
0.500 |
0.139 |
| 277 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
0.498 |
0.075 |
| 278 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.494 |
0.362 |
| 279 |
249977_at |
AT5G18820
|
Cpn60alpha2, chaperonin-60alpha2, EMB3007, embryo defective 3007 |
0.494 |
-0.011 |
| 280 |
245699_at |
AT5G04250
|
[Cysteine proteinases superfamily protein] |
0.494 |
0.155 |
| 281 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
0.493 |
0.090 |
| 282 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.493 |
0.430 |
| 283 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
0.493 |
0.112 |
| 284 |
253219_at |
AT4G34990
|
AtMYB32, myb domain protein 32, MYB32, myb domain protein 32 |
0.491 |
0.156 |
| 285 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
0.489 |
0.069 |
| 286 |
264357_at |
AT1G03360
|
ATRRP4, ribosomal RNA processing 4, RRP4, ribosomal RNA processing 4 |
0.489 |
-0.009 |
| 287 |
253092_at |
AT4G37380
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.488 |
0.017 |
| 288 |
263829_at |
AT2G40435
|
unknown |
0.488 |
0.032 |
| 289 |
262014_at |
AT1G35660
|
unknown |
0.487 |
0.089 |
| 290 |
251915_at |
AT3G53940
|
[Mitochondrial substrate carrier family protein] |
0.486 |
-0.019 |
| 291 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.485 |
0.381 |
| 292 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
0.483 |
-0.038 |
| 293 |
266002_at |
AT2G37310
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.481 |
-0.048 |
| 294 |
267576_at |
AT2G30640
|
MUG2, MUSTANG 2 |
0.481 |
0.284 |
| 295 |
254190_at |
AT4G23885
|
unknown |
0.480 |
-0.055 |
| 296 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.480 |
0.269 |
| 297 |
262124_at |
AT1G59660
|
[Nucleoporin autopeptidase] |
0.480 |
0.254 |
| 298 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
0.478 |
0.127 |
| 299 |
264251_at |
AT1G09190
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.477 |
-0.019 |
| 300 |
244911_at |
ATMG00820
|
ORF170 |
0.477 |
0.048 |
| 301 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-1.479 |
0.030 |
| 302 |
256527_at |
AT1G66100
|
[Plant thionin] |
-1.417 |
0.013 |
| 303 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-1.277 |
-0.016 |
| 304 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-1.171 |
-0.082 |
| 305 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-1.156 |
-0.026 |
| 306 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-1.148 |
0.117 |
| 307 |
247474_at |
AT5G62280
|
unknown |
-1.134 |
-0.305 |
| 308 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-1.133 |
0.038 |
| 309 |
258225_at |
AT3G15630
|
unknown |
-1.127 |
0.036 |
| 310 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-1.111 |
-0.092 |
| 311 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-1.109 |
0.028 |
| 312 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.101 |
0.074 |
| 313 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-1.077 |
0.082 |
| 314 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
-1.050 |
0.275 |
| 315 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-1.045 |
0.023 |
| 316 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
-1.041 |
-0.135 |
| 317 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-1.041 |
-0.054 |
| 318 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-1.032 |
-0.171 |
| 319 |
247754_at |
AT5G59080
|
unknown |
-1.031 |
-0.042 |
| 320 |
247878_at |
AT5G57760
|
unknown |
-1.028 |
0.022 |
| 321 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-1.027 |
-0.126 |
| 322 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-1.006 |
-0.050 |
| 323 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.991 |
0.005 |
| 324 |
265075_at |
AT1G55450
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.981 |
0.087 |
| 325 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-0.971 |
-0.026 |
| 326 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.970 |
0.047 |
| 327 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-0.960 |
0.176 |
| 328 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.956 |
0.018 |
| 329 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.945 |
-0.009 |
| 330 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.940 |
0.044 |
| 331 |
264774_at |
AT1G22890
|
unknown |
-0.938 |
0.013 |
| 332 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.932 |
0.019 |
| 333 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
-0.922 |
0.041 |
| 334 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.918 |
0.022 |
| 335 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
-0.912 |
-0.009 |
| 336 |
266658_at |
AT2G25735
|
unknown |
-0.905 |
-0.099 |
| 337 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.903 |
-0.152 |
| 338 |
244939_at |
ATCG00065
|
RPS12, RIBOSOMAL PROTEIN S12, RPS12A, ribosomal protein S12A |
-0.894 |
0.053 |
| 339 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.886 |
0.022 |
| 340 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
-0.884 |
-0.146 |
| 341 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
-0.881 |
-0.000 |
| 342 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.879 |
-0.052 |
| 343 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.872 |
0.006 |
| 344 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.866 |
0.001 |
| 345 |
265539_at |
AT2G15830
|
unknown |
-0.865 |
0.217 |
| 346 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
-0.862 |
0.067 |
| 347 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.859 |
-0.225 |
| 348 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.852 |
0.045 |
| 349 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.851 |
0.052 |
| 350 |
255904_at |
AT1G17860
|
[Kunitz family trypsin and protease inhibitor protein] |
-0.848 |
0.176 |
| 351 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
-0.848 |
0.054 |
| 352 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.845 |
0.318 |
| 353 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.842 |
-0.261 |
| 354 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.838 |
-0.103 |
| 355 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.837 |
-0.202 |
| 356 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.833 |
0.080 |
| 357 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.828 |
0.056 |
| 358 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.825 |
0.026 |
| 359 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.822 |
0.030 |
| 360 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
-0.819 |
-0.116 |
| 361 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
-0.817 |
-0.121 |
| 362 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.815 |
-0.000 |
| 363 |
257673_at |
AT3G20370
|
[TRAF-like family protein] |
-0.809 |
-0.129 |
| 364 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.808 |
0.378 |
| 365 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
-0.804 |
-0.043 |
| 366 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.804 |
0.031 |
| 367 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.804 |
-0.036 |
| 368 |
249242_at |
AT5G42250
|
[Zinc-binding alcohol dehydrogenase family protein] |
-0.802 |
-0.038 |
| 369 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
-0.800 |
0.023 |
| 370 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.795 |
-0.175 |
| 371 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
-0.791 |
0.055 |
| 372 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.789 |
-0.073 |
| 373 |
246270_at |
AT4G36500
|
unknown |
-0.787 |
0.045 |
| 374 |
261254_at |
AT1G05805
|
AKS2, ABA-responsive kinase substrate 2 |
-0.786 |
-0.126 |
| 375 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
-0.785 |
-0.064 |
| 376 |
249346_at |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
-0.783 |
0.061 |
| 377 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.782 |
0.049 |
| 378 |
265387_at |
AT2G20670
|
unknown |
-0.781 |
-0.029 |
| 379 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-0.780 |
-0.015 |
| 380 |
246565_at |
AT5G15530
|
BCCP2, biotin carboxyl carrier protein 2, CAC1-B |
-0.778 |
-0.068 |
| 381 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
-0.778 |
0.081 |
| 382 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.777 |
0.115 |
| 383 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
-0.774 |
-0.303 |
| 384 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
-0.770 |
0.015 |
| 385 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
-0.764 |
-0.149 |
| 386 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
-0.763 |
0.036 |
| 387 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.757 |
0.074 |
| 388 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
-0.757 |
0.076 |
| 389 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.752 |
0.074 |
| 390 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.749 |
-0.027 |
| 391 |
247444_at |
AT5G62630
|
HIPL2, hipl2 protein precursor |
-0.747 |
0.002 |
| 392 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.747 |
-0.226 |
| 393 |
246929_at |
AT5G25210
|
unknown |
-0.741 |
0.059 |
| 394 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.737 |
0.032 |
| 395 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
-0.737 |
-0.231 |
| 396 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.729 |
0.051 |
| 397 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
-0.727 |
-0.102 |
| 398 |
251036_at |
AT5G02160
|
unknown |
-0.727 |
0.076 |
| 399 |
259791_at |
AT1G29700
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
-0.726 |
-0.003 |
| 400 |
256091_at |
AT1G20693
|
HMG BETA 1, HIGH MOBILITY GROUP BETA 1, HMGB2, high mobility group B2, NFD02, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP D 02, NFD2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP D 2 |
-0.724 |
-0.252 |
| 401 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
-0.723 |
-0.111 |
| 402 |
249726_at |
AT5G35480
|
unknown |
-0.722 |
0.046 |
| 403 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.722 |
0.025 |
| 404 |
245692_at |
AT5G04150
|
BHLH101 |
-0.721 |
0.024 |
| 405 |
261221_at |
AT1G19960
|
unknown |
-0.719 |
-0.003 |
| 406 |
260975_at |
AT1G53430
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.719 |
-0.077 |
| 407 |
246487_at |
AT5G16030
|
unknown |
-0.716 |
0.021 |
| 408 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.716 |
-0.340 |
| 409 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.712 |
-0.284 |
| 410 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
-0.712 |
0.057 |
| 411 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
-0.711 |
0.001 |
| 412 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.710 |
0.160 |
| 413 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
-0.709 |
-0.629 |
| 414 |
259327_at |
AT3G16460
|
JAL34, jacalin-related lectin 34 |
-0.707 |
-0.092 |
| 415 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.703 |
0.031 |
| 416 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.702 |
0.016 |
| 417 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.700 |
0.099 |
| 418 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.698 |
0.003 |
| 419 |
256469_at |
AT1G32540
|
LOL1, lsd one like 1 |
-0.698 |
0.030 |
| 420 |
256170_at |
AT1G51790
|
[Leucine-rich repeat protein kinase family protein] |
-0.697 |
0.014 |
| 421 |
249230_at |
AT5G42070
|
unknown |
-0.692 |
0.026 |
| 422 |
248509_at |
AT5G50335
|
unknown |
-0.692 |
0.066 |
| 423 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
-0.688 |
-0.066 |
| 424 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.688 |
-0.106 |
| 425 |
257381_at |
AT2G37950
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.686 |
-0.112 |
| 426 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
-0.685 |
0.045 |
| 427 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
-0.684 |
0.009 |
| 428 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.683 |
-0.007 |
| 429 |
266516_at |
AT2G47880
|
[Glutaredoxin family protein] |
-0.680 |
-0.076 |
| 430 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
-0.680 |
0.031 |
| 431 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.680 |
-0.152 |
| 432 |
267580_at |
AT2G41990
|
unknown |
-0.678 |
-0.060 |
| 433 |
263558_at |
AT2G16380
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.678 |
0.066 |
| 434 |
249718_at |
AT5G35740
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.675 |
-0.142 |
| 435 |
248942_at |
AT5G45480
|
unknown |
-0.669 |
-0.145 |
| 436 |
258468_at |
AT3G06070
|
unknown |
-0.669 |
-0.148 |
| 437 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.669 |
0.011 |
| 438 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.668 |
0.029 |
| 439 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.668 |
0.081 |
| 440 |
265611_at |
AT2G25510
|
unknown |
-0.667 |
0.067 |
| 441 |
262899_at |
AT1G59870
|
ABCG36, ATP-binding cassette G36, ATABCG36, Arabidopsis thaliana ATP-binding cassette G36, ATPDR8, ARABIDOPSIS PLEIOTROPIC DRUG RESISTANCE 8, PDR8, PLEIOTROPIC DRUG RESISTANCE 8, PEN3, PENETRATION 3 |
-0.662 |
0.030 |
| 442 |
264379_at |
AT2G25200
|
unknown |
-0.661 |
-0.057 |
| 443 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.660 |
-0.067 |
| 444 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
-0.660 |
0.002 |
| 445 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
-0.659 |
0.097 |
| 446 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
-0.658 |
-0.193 |
| 447 |
252976_s_at |
AT4G38550
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.657 |
0.009 |
| 448 |
249378_at |
AT5G40450
|
unknown |
-0.657 |
-0.123 |
| 449 |
246997_at |
AT5G67390
|
unknown |
-0.654 |
-0.005 |
| 450 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.653 |
-0.051 |
| 451 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.653 |
-0.069 |
| 452 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.651 |
-0.042 |
| 453 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.645 |
-0.062 |
| 454 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.643 |
-0.022 |
| 455 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
-0.642 |
-0.188 |
| 456 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.642 |
-0.788 |
| 457 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
-0.640 |
-0.061 |
| 458 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
-0.638 |
-0.123 |
| 459 |
262096_at |
AT1G56010
|
anac021, Arabidopsis NAC domain containing protein 21, ANAC022, Arabidopsis NAC domain containing protein 22, NAC1, NAC domain containing protein 1 |
-0.638 |
0.007 |
| 460 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.637 |
0.040 |
| 461 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.635 |
-0.033 |
| 462 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-0.635 |
0.270 |
| 463 |
245117_at |
AT2G41560
|
ACA4, autoinhibited Ca(2+)-ATPase, isoform 4 |
-0.634 |
-0.022 |
| 464 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.634 |
0.240 |
| 465 |
249887_at |
AT5G22310
|
unknown |
-0.633 |
-0.079 |
| 466 |
254954_at |
AT4G10910
|
unknown |
-0.632 |
0.042 |
| 467 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
-0.632 |
0.036 |
| 468 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
-0.631 |
0.036 |
| 469 |
260472_at |
AT1G10990
|
unknown |
-0.630 |
0.018 |
| 470 |
249919_at |
AT5G19250
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.630 |
-0.025 |
| 471 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.629 |
-0.038 |
| 472 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
-0.628 |
-0.018 |
| 473 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
-0.625 |
0.235 |
| 474 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.625 |
-0.300 |
| 475 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
-0.624 |
-0.130 |
| 476 |
245930_at |
AT5G09240
|
[ssDNA-binding transcriptional regulator] |
-0.623 |
-0.067 |
| 477 |
257191_at |
AT3G13175
|
unknown |
-0.619 |
0.069 |
| 478 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
-0.618 |
-0.039 |
| 479 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
-0.618 |
-0.219 |
| 480 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.615 |
-0.088 |
| 481 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.610 |
0.016 |
| 482 |
259996_at |
AT1G67910
|
unknown |
-0.608 |
-0.293 |
| 483 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.606 |
-0.038 |
| 484 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
-0.605 |
0.245 |
| 485 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.605 |
-0.053 |
| 486 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
-0.604 |
0.052 |
| 487 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.603 |
-0.267 |
| 488 |
260668_at |
AT1G19530
|
unknown |
-0.601 |
0.232 |
| 489 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
-0.600 |
-0.088 |
| 490 |
266335_at |
AT2G32440
|
ATKAO2, ARABIDOPSIS ENT-KAURENOIC ACID HYDROXYLASE 2, CYP88A4, KAO2, ent-kaurenoic acid hydroxylase 2 |
-0.597 |
0.076 |
| 491 |
262772_at |
AT1G13210
|
ACA.l, autoinhibited Ca2+/ATPase II |
-0.594 |
-0.034 |
| 492 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.591 |
-0.115 |
| 493 |
262700_at |
AT1G76020
|
[Thioredoxin superfamily protein] |
-0.590 |
0.013 |
| 494 |
251142_at |
AT5G01015
|
unknown |
-0.590 |
0.089 |
| 495 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.589 |
-0.154 |
| 496 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.589 |
0.034 |
| 497 |
261175_at |
AT1G04800
|
[glycine-rich protein] |
-0.588 |
0.089 |
| 498 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
-0.587 |
0.015 |
| 499 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
-0.586 |
-0.110 |
| 500 |
252511_at |
AT3G46280
|
[protein kinase-related] |
-0.585 |
-0.039 |
| 501 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
-0.585 |
0.496 |
| 502 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-0.584 |
0.231 |
| 503 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
-0.580 |
0.296 |
| 504 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.579 |
-0.177 |
| 505 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.579 |
-0.248 |
| 506 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
-0.579 |
-0.097 |
| 507 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.578 |
0.023 |
| 508 |
256796_at |
AT3G22210
|
unknown |
-0.578 |
-0.074 |
| 509 |
260083_at |
AT1G63220
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.577 |
-0.253 |
| 510 |
265893_at |
AT2G15042
|
[Leucine-rich repeat (LRR) family protein] |
-0.577 |
0.044 |
| 511 |
260877_at |
AT1G21500
|
unknown |
-0.574 |
0.036 |
| 512 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.572 |
-0.100 |
| 513 |
249742_at |
AT5G24490
|
[30S ribosomal protein, putative] |
-0.571 |
0.102 |
| 514 |
259166_at |
AT3G01670
|
AtSEOR2, Arabidopsis thaliana sieve element occlusion-related 2, SEOa, sieve element occlusion a, SEOR2, sieve element occlusion-related 2 |
-0.570 |
-0.045 |
| 515 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.569 |
0.004 |
| 516 |
254919_at |
AT4G11360
|
RHA1B, RING-H2 finger A1B |
-0.569 |
0.122 |
| 517 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.568 |
0.011 |
| 518 |
257076_at |
AT3G19680
|
unknown |
-0.567 |
-0.302 |
| 519 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.566 |
0.038 |
| 520 |
266552_at |
AT2G46330
|
AGP16, arabinogalactan protein 16, ATAGP16 |
-0.566 |
-0.059 |
| 521 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.566 |
-0.071 |
| 522 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.565 |
0.051 |
| 523 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.565 |
-0.005 |
| 524 |
245690_at |
AT5G04230
|
ATPAL3, PAL3, phenyl alanine ammonia-lyase 3 |
-0.563 |
-0.064 |
| 525 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
-0.563 |
0.028 |
| 526 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.563 |
0.175 |
| 527 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.562 |
-0.308 |
| 528 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
-0.561 |
0.024 |
| 529 |
250810_at |
AT5G05090
|
[Homeodomain-like superfamily protein] |
-0.561 |
-0.069 |
| 530 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.561 |
-0.149 |
| 531 |
264096_at |
AT1G78995
|
unknown |
-0.560 |
0.073 |
| 532 |
263953_at |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
-0.559 |
-0.011 |
| 533 |
267636_at |
AT2G42110
|
unknown |
-0.558 |
-0.232 |
| 534 |
260921_at |
AT1G21540
|
[AMP-dependent synthetase and ligase family protein] |
-0.557 |
-0.016 |
| 535 |
259373_at |
AT1G69160
|
unknown |
-0.557 |
0.009 |
| 536 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-0.557 |
0.024 |
| 537 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
-0.556 |
0.049 |
| 538 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.555 |
-0.328 |
| 539 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.555 |
-0.455 |
| 540 |
261594_at |
AT1G33240
|
AT-GTL1, GT2-LIKE 1, ATGTL1, GTL1, GT-2-like 1 |
-0.554 |
-0.277 |
| 541 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
-0.553 |
0.079 |
| 542 |
254251_at |
AT4G23300
|
CRK22, cysteine-rich RLK (RECEPTOR-like protein kinase) 22 |
-0.551 |
0.000 |
| 543 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.549 |
0.076 |
| 544 |
252906_at |
AT4G39640
|
GGT1, gamma-glutamyl transpeptidase 1 |
-0.548 |
0.021 |
| 545 |
245353_at |
AT4G16000
|
unknown |
-0.546 |
0.045 |
| 546 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.546 |
-0.204 |
| 547 |
267209_at |
AT2G30930
|
unknown |
-0.542 |
-0.418 |
| 548 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
-0.540 |
0.028 |
| 549 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
-0.537 |
-0.123 |
| 550 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.537 |
0.061 |
| 551 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
-0.537 |
0.065 |
| 552 |
255148_at |
AT4G08470
|
MAPKKK10, MEKK3, MAPK/ERK kinase kinase 3 |
-0.536 |
-0.017 |
| 553 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.535 |
-0.072 |
| 554 |
248044_at |
AT5G56020
|
[Got1/Sft2-like vescicle transport protein family] |
-0.534 |
0.033 |
| 555 |
264998_at |
AT1G67330
|
unknown |
-0.534 |
-0.169 |
| 556 |
260974_at |
AT1G53440
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.533 |
-0.015 |
| 557 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.532 |
-0.114 |
| 558 |
251684_at |
AT3G56410
|
unknown |
-0.532 |
-0.034 |
| 559 |
251816_at |
AT3G55005
|
TON1B, tonneau 1b |
-0.532 |
-0.059 |
| 560 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.530 |
-0.038 |
| 561 |
257798_at |
AT3G15950
|
NAI2 |
-0.530 |
-0.129 |
| 562 |
264238_at |
AT1G54740
|
unknown |
-0.529 |
-0.190 |
| 563 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.528 |
-0.159 |
| 564 |
250547_at |
AT5G08100
|
ASPGA1, asparaginase A1 |
-0.526 |
-0.048 |
| 565 |
254329_at |
AT4G22540
|
ORP2A, OSBP(oxysterol binding protein)-related protein 2A |
-0.526 |
-0.036 |
| 566 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
-0.526 |
-0.018 |
| 567 |
249485_at |
AT5G39020
|
[Malectin/receptor-like protein kinase family protein] |
-0.526 |
0.043 |
| 568 |
254578_at |
AT4G19410
|
[Pectinacetylesterase family protein] |
-0.526 |
-0.007 |
| 569 |
256569_at |
AT3G19550
|
unknown |
-0.523 |
-0.004 |
| 570 |
250603_at |
AT5G07820
|
[Plant calmodulin-binding protein-related] |
-0.523 |
-0.142 |
| 571 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.523 |
-0.687 |
| 572 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-0.522 |
0.282 |
| 573 |
259181_at |
AT3G01690
|
[alpha/beta-Hydrolases superfamily protein] |
-0.522 |
-0.048 |
| 574 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.521 |
-0.038 |
| 575 |
267289_at |
AT2G23770
|
LYK4, LysM-containing receptor-like kinase 4 |
-0.521 |
0.257 |
| 576 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.521 |
0.001 |
| 577 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
-0.520 |
-0.265 |
| 578 |
247087_at |
AT5G66330
|
[Leucine-rich repeat (LRR) family protein] |
-0.520 |
-0.164 |
| 579 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.519 |
-0.152 |
| 580 |
261492_at |
AT1G14290
|
SBH2, sphingoid base hydroxylase 2 |
-0.519 |
-0.114 |
| 581 |
246366_at |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
-0.519 |
-0.146 |
| 582 |
258530_at |
AT3G06840
|
unknown |
-0.519 |
0.060 |
| 583 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.518 |
-0.022 |
| 584 |
267381_at |
AT2G26190
|
[calmodulin-binding family protein] |
-0.516 |
-0.011 |
| 585 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
-0.516 |
-0.053 |
| 586 |
264133_at |
AT1G79080
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.516 |
-0.116 |
| 587 |
260167_at |
AT1G71970
|
unknown |
-0.516 |
-0.185 |
| 588 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.514 |
-0.127 |
| 589 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
-0.513 |
-0.061 |
| 590 |
252053_at |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
-0.512 |
-0.003 |
| 591 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
-0.510 |
0.124 |
| 592 |
256455_at |
AT1G75190
|
unknown |
-0.510 |
0.004 |
| 593 |
254561_at |
AT4G19160
|
unknown |
-0.509 |
-0.091 |
| 594 |
256149_at |
AT1G55110
|
AtIDD7, indeterminate(ID)-domain 7, IDD7, indeterminate(ID)-domain 7 |
-0.509 |
0.089 |
| 595 |
257939_at |
AT3G19930
|
ATSTP4, SUGAR TRANSPORTER 4, STP4, sugar transporter 4 |
-0.509 |
-0.077 |
| 596 |
255596_at |
AT4G01720
|
AtWRKY47, WRKY47 |
-0.509 |
-0.169 |
| 597 |
266500_at |
AT2G06925
|
ATSPLA2-ALPHA, PHOSPHOLIPASE A2-ALPHA, PLA2-ALPHA |
-0.509 |
-0.149 |
| 598 |
258617_at |
AT3G03000
|
[EF hand calcium-binding protein family] |
-0.509 |
-0.030 |
| 599 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.508 |
-0.121 |
| 600 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.508 |
-0.238 |