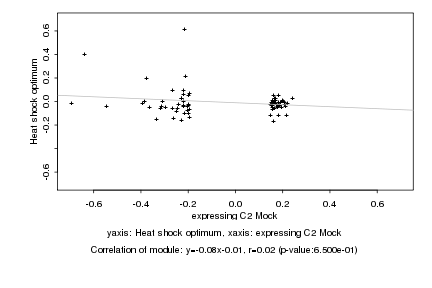
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
expressing C2 Mock |
Log2 signal ratio
Heat shock optimum |
| 1 |
244931_at |
ATMG00630
|
ORF110B |
0.241 |
0.032 |
| 2 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.218 |
-0.013 |
| 3 |
253166_at |
AT4G35290
|
ATGLR3.2, ATGLUR2, GLR3.2, GLUTAMATE RECEPTOR 3.2, GLUR2, glutamate receptor 2 |
0.216 |
-0.111 |
| 4 |
250322_at |
AT5G12870
|
ATMYB46, MYB DOMAIN PROTEIN 46, MYB46, myb domain protein 46 |
0.209 |
-0.037 |
| 5 |
255530_at |
AT4G02140
|
unknown |
0.207 |
-0.017 |
| 6 |
267572_at |
AT2G30660
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.206 |
-0.007 |
| 7 |
266689_at |
AT2G19930
|
[RNA-dependent RNA polymerase family protein] |
0.200 |
0.008 |
| 8 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.197 |
0.006 |
| 9 |
257402_at |
AT1G23570
|
unknown |
0.195 |
0.014 |
| 10 |
244949_at |
ATMG00150
|
ORF116 |
0.192 |
-0.050 |
| 11 |
247903_at |
AT5G57340
|
unknown |
0.188 |
-0.000 |
| 12 |
264496_at |
AT1G27285
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:AAC02666 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
0.186 |
-0.026 |
| 13 |
263546_at |
AT2G21550
|
[Bifunctional dihydrofolate reductase/thymidylate synthase] |
0.183 |
-0.032 |
| 14 |
266038_at |
AT2G07680
|
ABCC13, ATP-binding cassette C13, ATMRP11, multidrug resistance-associated protein 11, MRP11, multidrug resistance-associated protein 11 |
0.182 |
-0.116 |
| 15 |
250227_at |
AT5G13830
|
[FtsJ-like methyltransferase family protein] |
0.181 |
-0.034 |
| 16 |
250468_at |
AT5G10120
|
[Ethylene insensitive 3 family protein] |
0.180 |
-0.002 |
| 17 |
265879_at |
AT2G42450
|
[alpha/beta-Hydrolases superfamily protein] |
0.180 |
0.054 |
| 18 |
245581_at |
AT4G14840
|
unknown |
0.177 |
-0.037 |
| 19 |
267573_at |
AT2G30670
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.177 |
-0.043 |
| 20 |
255640_at |
AT4G00800
|
SETH5 |
0.170 |
-0.019 |
| 21 |
254808_at |
AT4G12740
|
[HhH-GPD base excision DNA repair family protein] |
0.168 |
0.029 |
| 22 |
265492_at |
AT2G15660
|
AGL95, AGAMOUS-like 95 |
0.166 |
0.016 |
| 23 |
265997_at |
AT2G24250
|
unknown |
0.166 |
-0.014 |
| 24 |
253809_at |
AT4G28320
|
AtMAN5, MAN5, endo-beta-mannase 5 |
0.164 |
0.032 |
| 25 |
245129_at |
AT2G45350
|
CRR4, CHLORORESPIRATORY REDUCTION 4 |
0.164 |
0.017 |
| 26 |
260646_at |
AT1G53340
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.163 |
-0.054 |
| 27 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
0.160 |
-0.163 |
| 28 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.158 |
0.000 |
| 29 |
251147_at |
AT3G63480
|
[ATP binding microtubule motor family protein] |
0.157 |
0.059 |
| 30 |
245670_at |
AT1G28210
|
ATJ1 |
0.156 |
0.013 |
| 31 |
263108_at |
AT1G65240
|
[Eukaryotic aspartyl protease family protein] |
0.156 |
-0.012 |
| 32 |
254419_at |
AT4G21490
|
NDB3, NAD(P)H dehydrogenase B3 |
0.156 |
-0.063 |
| 33 |
248082_at |
AT5G55400
|
[Actin binding Calponin homology (CH) domain-containing protein] |
0.154 |
-0.048 |
| 34 |
253126_at |
AT4G36050
|
[endonuclease/exonuclease/phosphatase family protein] |
0.153 |
-0.033 |
| 35 |
250659_at |
AT5G07050
|
UMAMIT9, Usually multiple acids move in and out Transporters 9 |
0.152 |
-0.024 |
| 36 |
251583_at |
AT3G58590
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.152 |
-0.003 |
| 37 |
263587_at |
AT2G25340
|
ATVAMP712, vesicle-associated membrane protein 712, VAMP712, VAMP712, vesicle-associated membrane protein 712 |
0.149 |
-0.026 |
| 38 |
256325_at |
AT3G02330
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.147 |
-0.018 |
| 39 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
0.146 |
-0.111 |
| 40 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.697 |
-0.010 |
| 41 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.640 |
0.402 |
| 42 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
-0.549 |
-0.034 |
| 43 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.394 |
-0.013 |
| 44 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
-0.386 |
0.006 |
| 45 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.378 |
0.196 |
| 46 |
249729_at |
AT5G24410
|
PGL4, 6-phosphogluconolactonase 4 |
-0.364 |
-0.044 |
| 47 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.335 |
-0.148 |
| 48 |
255252_at |
AT4G04990
|
unknown |
-0.318 |
-0.059 |
| 49 |
266330_at |
AT2G01530
|
MLP329, MLP-like protein 329, ZCE2, (Zusammen-CA)-enhanced 2 |
-0.314 |
-0.037 |
| 50 |
263208_at |
AT1G10480
|
ZFP5, zinc finger protein 5 |
-0.311 |
0.005 |
| 51 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.297 |
-0.043 |
| 52 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.268 |
-0.053 |
| 53 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.268 |
0.094 |
| 54 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.265 |
-0.141 |
| 55 |
248237_at |
AT5G53890
|
AtPSKR2, PSKR2, phytosylfokine-alpha receptor 2 |
-0.252 |
-0.084 |
| 56 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.246 |
-0.059 |
| 57 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
-0.244 |
-0.020 |
| 58 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.231 |
-0.161 |
| 59 |
251284_at |
AT3G61840
|
unknown |
-0.231 |
0.032 |
| 60 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
-0.231 |
0.034 |
| 61 |
249721_at |
AT5G24655
|
LSU4, RESPONSE TO LOW SULFUR 4 |
-0.222 |
-0.028 |
| 62 |
252202_at |
AT3G50300
|
[HXXXD-type acyl-transferase family protein] |
-0.222 |
-0.038 |
| 63 |
249009_at |
AT5G44610
|
MAP18, microtubule-associated protein 18, PCAP2, PLASMA MEMBRANE ASSOCIATED CA2+-BINDING PROTEIN-2 |
-0.221 |
0.006 |
| 64 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
-0.221 |
0.102 |
| 65 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
-0.220 |
0.063 |
| 66 |
262560_at |
AT1G34280
|
unknown |
-0.218 |
-0.097 |
| 67 |
267459_at |
AT2G33850
|
unknown |
-0.217 |
0.616 |
| 68 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.214 |
0.213 |
| 69 |
256935_at |
AT3G22570
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.203 |
-0.071 |
| 70 |
252087_at |
AT3G52080
|
chx28, cation/hydrogen exchanger 28 |
-0.203 |
-0.039 |
| 71 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.202 |
-0.100 |
| 72 |
249252_at |
AT5G42010
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.201 |
-0.033 |
| 73 |
261381_at |
AT1G05460
|
SDE3, SILENCING DEFECTIVE |
-0.200 |
-0.023 |
| 74 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
-0.199 |
0.059 |
| 75 |
266760_at |
AT2G46870
|
NGA1, NGATHA1 |
-0.197 |
-0.134 |
| 76 |
251061_at |
AT5G01830
|
SAUR21, SMALL AUXIN UP RNA 21 |
-0.196 |
-0.067 |
| 77 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.196 |
0.075 |