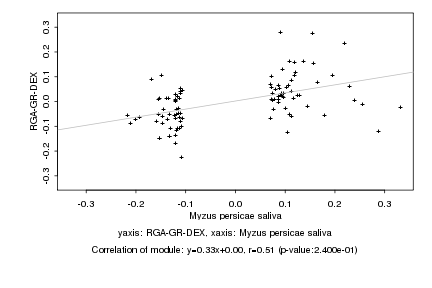
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Myzus persicae saliva |
Log2 signal ratio
RGA-GR-DEX |
| 1 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
0.331 |
-0.024 |
| 2 |
256789_at |
AT3G13672
|
SINA2 |
0.287 |
-0.119 |
| 3 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.255 |
-0.011 |
| 4 |
251012_at |
AT5G02580
|
unknown |
0.238 |
0.006 |
| 5 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.228 |
0.061 |
| 6 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.219 |
0.237 |
| 7 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
0.194 |
0.108 |
| 8 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
0.179 |
-0.053 |
| 9 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.164 |
0.077 |
| 10 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.155 |
0.155 |
| 11 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.153 |
0.277 |
| 12 |
264001_at |
AT2G22420
|
[Peroxidase superfamily protein] |
0.143 |
-0.017 |
| 13 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.136 |
0.163 |
| 14 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
0.128 |
0.027 |
| 15 |
266962_at |
AT2G39435
|
TRM18, TON1 Recruiting Motif 18 |
0.124 |
0.027 |
| 16 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.120 |
0.118 |
| 17 |
259856_at |
AT1G68440
|
unknown |
0.117 |
0.109 |
| 18 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
0.117 |
0.158 |
| 19 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.116 |
0.014 |
| 20 |
245217_at |
AT1G38340
|
[gypsy-like retrotransposon family (Athila), has a 3.7e-36 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
0.112 |
-0.058 |
| 21 |
254193_at |
AT4G23870
|
unknown |
0.111 |
0.087 |
| 22 |
246796_at |
AT5G26770
|
unknown |
0.111 |
0.041 |
| 23 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.108 |
0.163 |
| 24 |
255061_at |
AT4G08930
|
APRL6, APR-like 6, ATAPRL6, APR-like 6 |
0.107 |
-0.051 |
| 25 |
252173_at |
AT3G50650
|
[GRAS family transcription factor] |
0.105 |
0.067 |
| 26 |
264879_at |
AT1G61260
|
unknown |
0.104 |
-0.122 |
| 27 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.102 |
0.058 |
| 28 |
254014_at |
AT4G26120
|
[Ankyrin repeat family protein / BTB/POZ domain-containing protein] |
0.099 |
-0.026 |
| 29 |
263075_at |
AT2G17570
|
AtcPT1, cPT1, cis-prenyltransferase 1 |
0.096 |
0.020 |
| 30 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
0.095 |
0.036 |
| 31 |
249577_at |
AT5G37710
|
[alpha/beta-Hydrolases superfamily protein] |
0.094 |
0.132 |
| 32 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
0.093 |
0.024 |
| 33 |
248756_at |
AT5G47560
|
ATSDAT, ATTDT, TONOPLAST DICARBOXYLATE TRANSPORTER, TDT, tonoplast dicarboxylate transporter |
0.092 |
0.035 |
| 34 |
264910_at |
AT1G61100
|
[disease resistance protein (TIR class), putative] |
0.090 |
0.027 |
| 35 |
250580_at |
AT5G07440
|
GDH2, glutamate dehydrogenase 2 |
0.090 |
0.279 |
| 36 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.087 |
0.056 |
| 37 |
251379_at |
AT3G60680
|
unknown |
0.086 |
0.023 |
| 38 |
261973_at |
AT1G64610
|
[Transducin/WD40 repeat-like superfamily protein] |
0.085 |
0.011 |
| 39 |
267532_at |
AT2G42040
|
unknown |
0.085 |
-0.003 |
| 40 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
0.085 |
0.067 |
| 41 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.079 |
0.049 |
| 42 |
245770_at |
AT1G30240
|
[FUNCTIONS IN: binding] |
0.078 |
0.009 |
| 43 |
253654_at |
AT4G30060
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.075 |
-0.030 |
| 44 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.074 |
0.007 |
| 45 |
257879_at |
AT3G17160
|
unknown |
0.073 |
0.036 |
| 46 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.072 |
0.102 |
| 47 |
261599_at |
AT1G49700
|
unknown |
0.071 |
0.010 |
| 48 |
250556_at |
AT5G07920
|
ATDGK1, DIACYLGLYCEROL KINASE 1, DGK1, diacylglycerol kinase1 |
0.071 |
0.058 |
| 49 |
257805_at |
AT3G18830
|
ATPLT5, POLYOL TRANSPORTER 5, ATPMT5, ARABIDOPSIS THALIANA POLYOL/MONOSACCHARIDE TRANSPORTER 5, PMT5, polyol/monosaccharide transporter 5 |
0.070 |
0.071 |
| 50 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.069 |
-0.066 |
| 51 |
260112_at |
AT1G63310
|
unknown |
-0.217 |
-0.055 |
| 52 |
258807_at |
AT3G04030
|
MYR2 |
-0.211 |
-0.086 |
| 53 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.202 |
-0.071 |
| 54 |
264184_at |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.193 |
-0.062 |
| 55 |
261256_at |
AT1G05760
|
RTM1, restricted tev movement 1 |
-0.170 |
0.090 |
| 56 |
263136_at |
AT1G78580
|
ATTPS1, trehalose-6-phosphate synthase, TPS1, trehalose-6-phosphate synthase, TPS1, TREHALOSE-6-PHOSPHATE SYNTHASE 1 |
-0.159 |
-0.080 |
| 57 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.156 |
-0.052 |
| 58 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.155 |
0.010 |
| 59 |
262120_at |
AT1G02950
|
ATGSTF4, glutathione S-transferase F4, GST31, GLUTATHIONE S-TRANSFERASE 31, GSTF4, glutathione S-transferase F4 |
-0.154 |
0.014 |
| 60 |
255825_at |
AT2G40475
|
ASG8, ALTERED SEED GERMINATION 8 |
-0.153 |
-0.149 |
| 61 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.153 |
-0.147 |
| 62 |
259166_at |
AT3G01670
|
AtSEOR2, Arabidopsis thaliana sieve element occlusion-related 2, SEOa, sieve element occlusion a, SEOR2, sieve element occlusion-related 2 |
-0.150 |
0.107 |
| 63 |
257081_at |
AT3G30460
|
[RING/U-box superfamily protein] |
-0.148 |
-0.060 |
| 64 |
258414_at |
AT3G17380
|
[TRAF-like family protein] |
-0.148 |
-0.088 |
| 65 |
249214_at |
AT5G42720
|
[Glycosyl hydrolase family 17 protein] |
-0.145 |
-0.032 |
| 66 |
254629_at |
AT4G18425
|
unknown |
-0.139 |
0.014 |
| 67 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
-0.137 |
-0.072 |
| 68 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.135 |
0.013 |
| 69 |
251142_at |
AT5G01015
|
unknown |
-0.134 |
-0.139 |
| 70 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
-0.134 |
-0.139 |
| 71 |
253754_at |
AT4G29020
|
[glycine-rich protein] |
-0.133 |
-0.051 |
| 72 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.131 |
-0.105 |
| 73 |
267465_at |
AT2G19170
|
SLP3, subtilisin-like serine protease 3 |
-0.123 |
-0.055 |
| 74 |
248210_at |
AT5G54000
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.122 |
0.008 |
| 75 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
-0.122 |
0.032 |
| 76 |
252503_at |
AT3G46910
|
[Cullin family protein] |
-0.122 |
-0.067 |
| 77 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.121 |
-0.166 |
| 78 |
259371_at |
AT1G69080
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.121 |
0.012 |
| 79 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.121 |
-0.137 |
| 80 |
245849_at |
AT5G13520
|
[peptidase M1 family protein] |
-0.121 |
0.001 |
| 81 |
262533_at |
AT1G17090
|
unknown |
-0.120 |
-0.049 |
| 82 |
251058_at |
AT5G01790
|
unknown |
-0.120 |
-0.113 |
| 83 |
260491_at |
AT1G51440
|
DALL2, DAD1-Like Lipase 2 |
-0.119 |
-0.031 |
| 84 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
-0.118 |
-0.108 |
| 85 |
249958_at |
AT5G18970
|
[AWPM-19-like family protein] |
-0.117 |
0.024 |
| 86 |
257093_at |
AT3G20570
|
AtENODL9, ENODL9, early nodulin-like protein 9 |
-0.116 |
-0.048 |
| 87 |
262039_at |
AT1G80050
|
APT2, adenine phosphoribosyl transferase 2, ATAPT2, PHT1.1 |
-0.115 |
-0.027 |
| 88 |
262154_at |
AT1G52700
|
[alpha/beta-Hydrolases superfamily protein] |
-0.114 |
-0.106 |
| 89 |
265250_at |
AT2G01950
|
BRL2, BRI1-like 2, VH1, VASCULAR HIGHWAY 1 |
-0.114 |
0.014 |
| 90 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.113 |
-0.063 |
| 91 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.112 |
0.041 |
| 92 |
258122_at |
AT3G14570
|
ATGSL04, glucan synthase-like 4, atgsl4, gsl04, GSL04, glucan synthase-like 4 |
-0.112 |
0.055 |
| 93 |
252010_at |
AT3G52740
|
unknown |
-0.111 |
0.033 |
| 94 |
265892_at |
AT2G15020
|
unknown |
-0.111 |
-0.077 |
| 95 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
-0.111 |
-0.046 |
| 96 |
267414_at |
AT2G34790
|
EDA28, EMBRYO SAC DEVELOPMENT ARREST 28, MEE23, MATERNAL EFFECT EMBRYO ARREST 23 |
-0.110 |
-0.099 |
| 97 |
248382_at |
AT5G51890
|
[Peroxidase superfamily protein] |
-0.109 |
-0.225 |
| 98 |
267284_at |
AT2G23700
|
unknown |
-0.107 |
0.046 |
| 99 |
246986_at |
AT5G67280
|
RLK, receptor-like kinase |
-0.107 |
-0.067 |