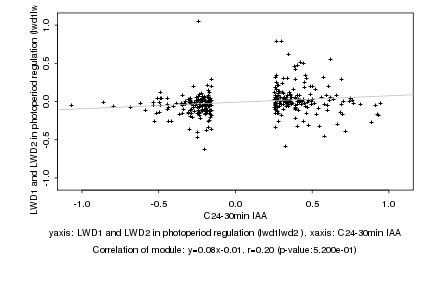
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
C24-30min IAA |
Log2 signal ratio
LWD1 and LWD2 in photoperiod regulation (lwd1lwd2 ) |
| 1 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
0.943 |
-0.023 |
| 2 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.930 |
-0.178 |
| 3 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
0.921 |
-0.158 |
| 4 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
0.908 |
-0.045 |
| 5 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
0.882 |
-0.266 |
| 6 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
0.814 |
-0.038 |
| 7 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.767 |
-0.020 |
| 8 |
266611_at |
AT2G14960
|
GH3.1 |
0.761 |
0.015 |
| 9 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.748 |
0.045 |
| 10 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
0.742 |
0.004 |
| 11 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.713 |
-0.381 |
| 12 |
245076_at |
AT2G23170
|
GH3.3 |
0.704 |
0.003 |
| 13 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
0.692 |
-0.161 |
| 14 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.691 |
0.292 |
| 15 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
0.687 |
-0.036 |
| 16 |
256190_at |
AT1G30100
|
ATNCED5, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 5, NCED5, nine-cis-epoxycarotenoid dioxygenase 5 |
0.679 |
-0.140 |
| 17 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
0.663 |
-0.291 |
| 18 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
0.652 |
0.085 |
| 19 |
247869_at |
AT5G57520
|
ATZFP2, ZINC FINGER PROTEIN 2, ZFP2, zinc finger protein 2 |
0.634 |
0.031 |
| 20 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
0.615 |
0.054 |
| 21 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.613 |
0.551 |
| 22 |
246798_at |
AT5G26930
|
GATA23, GATA transcription factor 23 |
0.612 |
0.024 |
| 23 |
263182_at |
AT1G05575
|
unknown |
0.606 |
0.208 |
| 24 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.599 |
-0.111 |
| 25 |
248316_at |
AT5G52670
|
[Copper transport protein family] |
0.596 |
-0.048 |
| 26 |
251178_at |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
0.590 |
0.087 |
| 27 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
0.576 |
-0.031 |
| 28 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
0.574 |
-0.454 |
| 29 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.570 |
0.320 |
| 30 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.559 |
0.059 |
| 31 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
0.544 |
-0.320 |
| 32 |
248801_at |
AT5G47370
|
HAT2 |
0.537 |
-0.083 |
| 33 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
0.525 |
-0.160 |
| 34 |
252765_at |
AT3G42800
|
unknown |
0.516 |
0.162 |
| 35 |
255945_at |
AT5G28610
|
unknown |
0.509 |
-0.014 |
| 36 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.502 |
0.029 |
| 37 |
261476_at |
AT1G14480
|
[Ankyrin repeat family protein] |
0.501 |
0.206 |
| 38 |
253179_at |
AT4G35200
|
unknown |
0.495 |
0.008 |
| 39 |
259027_at |
AT3G09280
|
unknown |
0.490 |
-0.029 |
| 40 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
0.487 |
0.203 |
| 41 |
251423_at |
AT3G60550
|
CYCP3;2, cyclin p3;2 |
0.486 |
0.092 |
| 42 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.471 |
-0.306 |
| 43 |
252103_at |
AT3G51410
|
unknown |
0.470 |
0.018 |
| 44 |
248278_at |
AT5G52890
|
[AT hook motif-containing protein] |
0.467 |
-0.072 |
| 45 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
0.465 |
-0.000 |
| 46 |
248789_at |
AT5G47440
|
unknown |
0.461 |
-0.148 |
| 47 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.459 |
0.308 |
| 48 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.456 |
-0.062 |
| 49 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
0.454 |
0.348 |
| 50 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.445 |
0.256 |
| 51 |
245041_at |
AT2G26530
|
AR781 |
0.445 |
0.188 |
| 52 |
254888_at |
AT4G11780
|
TRM10, TON1 Recruiting Motif 10 |
0.444 |
-0.013 |
| 53 |
247159_at |
AT5G65800
|
ACS5, ACC synthase 5, ATACS5, CIN5, CYTOKININ-INSENSITIVE 5, ETO2, ETHYLENE OVERPRODUCER 2 |
0.439 |
0.027 |
| 54 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.438 |
0.507 |
| 55 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
0.437 |
-0.250 |
| 56 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.432 |
0.003 |
| 57 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
0.430 |
-0.030 |
| 58 |
261777_at |
AT1G76210
|
unknown |
0.422 |
0.022 |
| 59 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.420 |
0.518 |
| 60 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.417 |
0.082 |
| 61 |
257026_at |
AT3G19200
|
unknown |
0.414 |
-0.006 |
| 62 |
255658_at |
AT4G00770
|
TRM9, TON1 Recruiting Motif 9 |
0.412 |
0.022 |
| 63 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.407 |
-0.111 |
| 64 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
0.407 |
0.038 |
| 65 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
0.402 |
0.081 |
| 66 |
262045_at |
AT1G80240
|
DGR1, DUF642 L-GalL responsive gene 1 |
0.402 |
0.005 |
| 67 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.401 |
0.473 |
| 68 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
0.399 |
-0.038 |
| 69 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.398 |
-0.322 |
| 70 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
0.391 |
-0.100 |
| 71 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.390 |
0.421 |
| 72 |
266908_at |
AT2G34650
|
ABR, ABRUPTUS, PID, PINOID |
0.388 |
0.298 |
| 73 |
247622_at |
AT5G60350
|
unknown |
0.388 |
-0.070 |
| 74 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.387 |
0.051 |
| 75 |
265144_at |
AT1G51170
|
AGC2-3, AGC2 kinase 3, UCN, UNICORN |
0.386 |
-0.049 |
| 76 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
0.386 |
-0.220 |
| 77 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.385 |
0.059 |
| 78 |
259785_at |
AT1G29490
|
SAUR68, SMALL AUXIN UPREGULATED 68 |
0.385 |
-0.082 |
| 79 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.381 |
0.459 |
| 80 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.375 |
0.164 |
| 81 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.371 |
-0.020 |
| 82 |
245108_at |
AT2G41510
|
ATCKX1, CYTOKININ OXIDASE/DEHYDROGENASE 1, CKX1, cytokinin oxidase/dehydrogenase 1 |
0.362 |
-0.036 |
| 83 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.360 |
-0.012 |
| 84 |
253827_at |
AT4G28085
|
unknown |
0.360 |
0.057 |
| 85 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.357 |
0.100 |
| 86 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
0.355 |
0.082 |
| 87 |
256872_at |
AT3G26490
|
[Phototropic-responsive NPH3 family protein] |
0.355 |
0.032 |
| 88 |
246992_at |
AT5G67430
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.354 |
0.009 |
| 89 |
263576_at |
AT2G17080
|
unknown |
0.353 |
-0.002 |
| 90 |
257832_at |
AT3G26740
|
CCL, CCR-like |
0.352 |
0.128 |
| 91 |
257386_at |
AT2G42440
|
ASL15, ASYMMETRIC LEAVES 2-like 15, LBD17, LOB domain-containing protein 17 |
0.350 |
0.003 |
| 92 |
251246_at |
AT3G62100
|
IAA30, indole-3-acetic acid inducible 30 |
0.347 |
0.012 |
| 93 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.345 |
-0.028 |
| 94 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.343 |
-0.005 |
| 95 |
257975_at |
AT3G20830
|
AGC2-4, AGC2 kinase 4, UCNL, UNICORN-like |
0.342 |
-0.061 |
| 96 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.340 |
0.627 |
| 97 |
253791_at |
AT4G28640
|
IAA11, indole-3-acetic acid inducible 11 |
0.339 |
-0.039 |
| 98 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
0.338 |
0.306 |
| 99 |
263931_at |
AT2G36220
|
unknown |
0.336 |
0.007 |
| 100 |
245209_at |
AT5G12340
|
unknown |
0.334 |
-0.016 |
| 101 |
253011_at |
AT4G37890
|
EDA40, embryo sac development arrest 40 |
0.333 |
0.023 |
| 102 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.333 |
0.079 |
| 103 |
247778_at |
AT5G58750
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.332 |
-0.060 |
| 104 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
0.330 |
0.005 |
| 105 |
248860_at |
AT5G46670
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.327 |
0.056 |
| 106 |
256583_at |
AT3G28850
|
[Glutaredoxin family protein] |
0.325 |
-0.010 |
| 107 |
247474_at |
AT5G62280
|
unknown |
0.321 |
-0.588 |
| 108 |
249533_at |
AT5G38790
|
unknown |
0.320 |
0.036 |
| 109 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
0.318 |
0.035 |
| 110 |
254962_at |
AT4G11050
|
AtGH9C3, glycosyl hydrolase 9C3, GH9C3, glycosyl hydrolase 9C3 |
0.317 |
0.026 |
| 111 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.314 |
0.058 |
| 112 |
258907_at |
AT3G06370
|
ATNHX4, NHX4, sodium hydrogen exchanger 4 |
0.312 |
0.117 |
| 113 |
267623_at |
AT2G39650
|
unknown |
0.312 |
0.308 |
| 114 |
264716_at |
AT1G70170
|
MMP, matrix metalloproteinase |
0.312 |
0.018 |
| 115 |
254408_at |
AT4G21390
|
B120 |
0.310 |
-0.094 |
| 116 |
266568_at |
AT2G24070
|
QWRF4, QWRF domain containing 4 |
0.310 |
0.041 |
| 117 |
260058_at |
AT1G78100
|
AUF1, auxin up-regulated f-box protein 1 |
0.309 |
-0.029 |
| 118 |
245755_at |
AT1G35210
|
unknown |
0.303 |
0.246 |
| 119 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.301 |
0.124 |
| 120 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
0.299 |
-0.036 |
| 121 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
0.297 |
-0.180 |
| 122 |
253643_at |
AT4G29780
|
unknown |
0.295 |
0.787 |
| 123 |
251374_at |
AT3G60390
|
HAT3, homeobox-leucine zipper protein 3 |
0.290 |
0.057 |
| 124 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
0.288 |
0.003 |
| 125 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.287 |
-0.019 |
| 126 |
261063_at |
AT1G07520
|
[GRAS family transcription factor] |
0.286 |
0.141 |
| 127 |
258253_at |
AT3G26760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.281 |
-0.065 |
| 128 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.280 |
0.221 |
| 129 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.279 |
0.083 |
| 130 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
0.279 |
-0.145 |
| 131 |
247047_at |
AT5G66650
|
unknown |
0.279 |
0.118 |
| 132 |
262105_at |
AT1G02810
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.278 |
0.114 |
| 133 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
0.277 |
-0.252 |
| 134 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.277 |
0.136 |
| 135 |
252053_at |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
0.275 |
0.135 |
| 136 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.273 |
0.070 |
| 137 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.273 |
0.221 |
| 138 |
265327_at |
AT2G18210
|
unknown |
0.272 |
-0.076 |
| 139 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
0.271 |
-0.152 |
| 140 |
253963_at |
AT4G26470
|
[Calcium-binding EF-hand family protein] |
0.271 |
0.014 |
| 141 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
0.270 |
0.211 |
| 142 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
0.269 |
0.039 |
| 143 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
0.268 |
-0.121 |
| 144 |
253044_at |
AT4G37290
|
unknown |
0.268 |
0.048 |
| 145 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
0.267 |
0.168 |
| 146 |
259274_at |
AT3G01220
|
ATHB20, homeobox protein 20, HB20, homeobox protein 20 |
0.266 |
-0.018 |
| 147 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.266 |
-0.049 |
| 148 |
250327_at |
AT5G12050
|
unknown |
0.265 |
-0.109 |
| 149 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.265 |
0.345 |
| 150 |
263379_at |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
0.264 |
0.257 |
| 151 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.263 |
0.792 |
| 152 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.261 |
0.317 |
| 153 |
251940_at |
AT3G53450
|
LOG4, LONELY GUY 4 |
0.261 |
-0.041 |
| 154 |
256010_at |
AT1G19220
|
ARF11, AUXIN RESPONSE FACTOR11, ARF19, auxin response factor 19, IAA22, indole-3-acetic acid inducible 22 |
0.261 |
-0.037 |
| 155 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.261 |
0.192 |
| 156 |
252129_at |
AT3G50890
|
AtHB28, homeobox protein 28, HB28, homeobox protein 28, ZHD7, ZINC FINGER HOMEODOMAIN 7 |
0.260 |
0.062 |
| 157 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
0.258 |
-0.009 |
| 158 |
258787_at |
AT3G11840
|
PUB24, plant U-box 24 |
0.258 |
-0.103 |
| 159 |
252204_at |
AT3G50340
|
unknown |
0.257 |
0.082 |
| 160 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
0.256 |
-0.329 |
| 161 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.256 |
-0.044 |
| 162 |
263002_at |
AT1G54200
|
unknown |
0.256 |
0.060 |
| 163 |
258388_at |
AT3G15370
|
ATEXP12, ATEXPA12, expansin 12, ATHEXP ALPHA 1.24, EXP12, EXPANSIN 12, EXPA12, expansin 12 |
0.255 |
-0.025 |
| 164 |
255028_at |
AT4G09890
|
unknown |
0.255 |
-0.149 |
| 165 |
248726_at |
AT5G47960
|
ATRABA4C, RAB GTPase homolog A4C, RABA4C, RAB GTPase homolog A4C, SMG1, SMALL MOLECULAR WEIGHT G-PROTEIN 1 |
0.254 |
0.126 |
| 166 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.254 |
-0.068 |
| 167 |
254063_at |
AT4G25390
|
[Protein kinase superfamily protein] |
0.253 |
0.038 |
| 168 |
266393_at |
AT2G41260
|
ATM17, M17 |
-1.073 |
-0.041 |
| 169 |
248125_at |
AT5G54740
|
SESA5, seed storage albumin 5 |
-0.862 |
-0.012 |
| 170 |
258327_at |
AT3G22640
|
PAP85 |
-0.801 |
-0.055 |
| 171 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.689 |
-0.066 |
| 172 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
-0.625 |
-0.025 |
| 173 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
-0.588 |
-0.107 |
| 174 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
-0.536 |
-0.012 |
| 175 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
-0.534 |
-0.051 |
| 176 |
265387_at |
AT2G20670
|
unknown |
-0.529 |
-0.257 |
| 177 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.519 |
-0.155 |
| 178 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.514 |
0.048 |
| 179 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
-0.501 |
-0.140 |
| 180 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
-0.499 |
-0.004 |
| 181 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
-0.492 |
-0.040 |
| 182 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.491 |
0.044 |
| 183 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
-0.491 |
0.122 |
| 184 |
261567_at |
AT1G33055
|
unknown |
-0.482 |
0.045 |
| 185 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
-0.449 |
0.047 |
| 186 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.449 |
-0.031 |
| 187 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.446 |
-0.095 |
| 188 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.442 |
-0.249 |
| 189 |
263881_at |
AT2G21820
|
unknown |
-0.441 |
-0.071 |
| 190 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.417 |
-0.258 |
| 191 |
247070_at |
AT5G66815
|
unknown |
-0.406 |
-0.063 |
| 192 |
254954_at |
AT4G10910
|
unknown |
-0.388 |
-0.024 |
| 193 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.371 |
-0.160 |
| 194 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.355 |
-0.022 |
| 195 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.348 |
-0.156 |
| 196 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
-0.348 |
-0.048 |
| 197 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.347 |
0.085 |
| 198 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.346 |
-0.028 |
| 199 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
-0.339 |
-0.103 |
| 200 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
-0.336 |
-0.019 |
| 201 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
-0.330 |
0.009 |
| 202 |
263208_at |
AT1G10480
|
ZFP5, zinc finger protein 5 |
-0.327 |
-0.026 |
| 203 |
246142_at |
AT5G19970
|
unknown |
-0.311 |
-0.152 |
| 204 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
-0.311 |
-0.134 |
| 205 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
-0.306 |
0.035 |
| 206 |
263970_at |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
-0.302 |
-0.009 |
| 207 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.302 |
-0.354 |
| 208 |
263138_at |
AT1G65090
|
unknown |
-0.302 |
0.035 |
| 209 |
245335_at |
AT4G16160
|
ATOEP16-2, ATOEP16-S |
-0.298 |
0.048 |
| 210 |
247585_at |
AT5G60680
|
unknown |
-0.294 |
0.023 |
| 211 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
-0.294 |
-0.113 |
| 212 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
-0.294 |
-0.004 |
| 213 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
-0.294 |
-0.008 |
| 214 |
247991_at |
AT5G56320
|
ATEXP14, ATEXPA14, expansin A14, ATHEXP ALPHA 1.5, EXP14, EXPANSIN 14, EXPA14, expansin A14 |
-0.293 |
-0.020 |
| 215 |
250173_at |
AT5G14340
|
AtMYB40, myb domain protein 40, MYB40, myb domain protein 40 |
-0.293 |
0.013 |
| 216 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
-0.293 |
-0.183 |
| 217 |
247754_at |
AT5G59080
|
unknown |
-0.292 |
-0.152 |
| 218 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-0.287 |
-0.099 |
| 219 |
249768_at |
AT5G24100
|
[Leucine-rich repeat protein kinase family protein] |
-0.287 |
-0.033 |
| 220 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.282 |
-0.201 |
| 221 |
265584_at |
AT2G20180
|
PIF1, PHY-INTERACTING FACTOR 1, PIL5, phytochrome interacting factor 3-like 5 |
-0.276 |
-0.073 |
| 222 |
259314_at |
AT3G05260
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.276 |
-0.044 |
| 223 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
-0.275 |
0.200 |
| 224 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.270 |
-0.130 |
| 225 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
-0.267 |
-0.008 |
| 226 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
-0.262 |
-0.045 |
| 227 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.257 |
-0.035 |
| 228 |
264640_at |
AT1G65680
|
ATEXPB2, expansin B2, ATHEXP BETA 1.4, EXPB2, expansin B2 |
-0.257 |
0.024 |
| 229 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
-0.255 |
0.059 |
| 230 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.252 |
-0.072 |
| 231 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.252 |
0.020 |
| 232 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
-0.251 |
-0.151 |
| 233 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
-0.250 |
-0.405 |
| 234 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-0.250 |
0.008 |
| 235 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.248 |
-0.467 |
| 236 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.247 |
0.021 |
| 237 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.245 |
0.070 |
| 238 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.245 |
-0.144 |
| 239 |
264379_at |
AT2G25200
|
unknown |
-0.244 |
-0.110 |
| 240 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
-0.243 |
0.072 |
| 241 |
265255_at |
AT2G28420
|
GLYI8, glyoxylase I 8 |
-0.242 |
0.031 |
| 242 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.242 |
1.053 |
| 243 |
247827_at |
AT5G58500
|
LSH5, LIGHT SENSITIVE HYPOCOTYLS 5 |
-0.239 |
-0.036 |
| 244 |
250103_at |
AT5G16600
|
AtMYB43, myb domain protein 43, MYB43, myb domain protein 43 |
-0.238 |
-0.056 |
| 245 |
253814_at |
AT4G28290
|
unknown |
-0.238 |
-0.173 |
| 246 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
-0.236 |
-0.146 |
| 247 |
253227_at |
AT4G35030
|
[Protein kinase superfamily protein] |
-0.235 |
-0.071 |
| 248 |
263175_at |
AT1G05510
|
OBAP1a, Oil Body-Associated Protein 1a |
-0.235 |
0.093 |
| 249 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
-0.235 |
-0.141 |
| 250 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.233 |
0.022 |
| 251 |
250937_at |
AT5G03230
|
unknown |
-0.229 |
-0.214 |
| 252 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
-0.229 |
-0.063 |
| 253 |
251857_at |
AT3G54770
|
ARP1, ABA-regulated RNA-binding protein 1 |
-0.228 |
-0.012 |
| 254 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
-0.227 |
0.115 |
| 255 |
255608_at |
AT4G01140
|
unknown |
-0.223 |
0.030 |
| 256 |
252511_at |
AT3G46280
|
[protein kinase-related] |
-0.221 |
-0.063 |
| 257 |
262863_at |
AT1G64910
|
[UDP-Glycosyltransferase superfamily protein] |
-0.221 |
0.071 |
| 258 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
-0.220 |
-0.129 |
| 259 |
258240_at |
AT3G27660
|
OLE3, OLEOSIN 3, OLEO4, oleosin 4 |
-0.220 |
0.031 |
| 260 |
251434_at |
AT3G59850
|
[Pectin lyase-like superfamily protein] |
-0.220 |
-0.044 |
| 261 |
259982_at |
AT1G76410
|
ATL8 |
-0.219 |
0.040 |
| 262 |
250472_at |
AT5G10210
|
unknown |
-0.219 |
-0.115 |
| 263 |
246911_at |
AT5G25810
|
tny, TINY |
-0.218 |
0.025 |
| 264 |
249937_at |
AT5G22470
|
[NAD+ ADP-ribosyltransferases] |
-0.218 |
0.050 |
| 265 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
-0.217 |
0.077 |
| 266 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
-0.217 |
-0.092 |
| 267 |
254333_at |
AT4G22753
|
ATSMO1-3, SMO1-3, sterol 4-alpha methyl oxidase 1-3 |
-0.215 |
0.046 |
| 268 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.215 |
0.005 |
| 269 |
264187_at |
AT1G54860
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.214 |
0.049 |
| 270 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
-0.214 |
0.027 |
| 271 |
251580_at |
AT3G58450
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.214 |
0.028 |
| 272 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
-0.210 |
-0.049 |
| 273 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.207 |
-0.615 |
| 274 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
-0.206 |
-0.037 |
| 275 |
262170_at |
AT1G74940
|
unknown |
-0.205 |
0.057 |
| 276 |
254175_at |
AT4G24050
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.205 |
0.069 |
| 277 |
266143_at |
AT2G38905
|
[Low temperature and salt responsive protein family] |
-0.202 |
0.035 |
| 278 |
249493_at |
AT5G39080
|
[HXXXD-type acyl-transferase family protein] |
-0.202 |
-0.093 |
| 279 |
259854_at |
AT1G72200
|
[RING/U-box superfamily protein] |
-0.201 |
-0.165 |
| 280 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
-0.200 |
-0.129 |
| 281 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
-0.199 |
0.006 |
| 282 |
257473_at |
AT1G33840
|
unknown |
-0.198 |
-0.035 |
| 283 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.198 |
-0.063 |
| 284 |
247522_at |
AT5G61340
|
unknown |
-0.197 |
-0.145 |
| 285 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.196 |
0.004 |
| 286 |
247483_at |
AT5G62420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.195 |
-0.021 |
| 287 |
259520_at |
AT1G12320
|
unknown |
-0.195 |
-0.045 |
| 288 |
252057_at |
AT3G52480
|
unknown |
-0.193 |
-0.022 |
| 289 |
246523_at |
AT5G15850
|
ATCOL1, BBX2, B-box domain protein 2, COL1, CONSTANS-like 1 |
-0.193 |
-0.375 |
| 290 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
-0.192 |
-0.086 |
| 291 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
-0.191 |
-0.047 |
| 292 |
246216_at |
AT4G36380
|
ROT3, ROTUNDIFOLIA 3 |
-0.189 |
-0.149 |
| 293 |
267579_at |
AT2G42000
|
AtMT4a, Arabidopsis thaliana metallothionein 4a |
-0.189 |
-0.035 |
| 294 |
254631_at |
AT4G18610
|
LSH9, LIGHT SENSITIVE HYPOCOTYLS 9 |
-0.188 |
0.059 |
| 295 |
267222_at |
AT2G43880
|
[Pectin lyase-like superfamily protein] |
-0.187 |
-0.052 |
| 296 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.186 |
0.222 |
| 297 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.185 |
-0.051 |
| 298 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
-0.184 |
-0.021 |
| 299 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
-0.183 |
-0.106 |
| 300 |
264210_at |
AT1G22640
|
ATMYB3, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 3, MYB3, myb domain protein 3 |
-0.183 |
-0.124 |
| 301 |
254440_at |
AT4G21020
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.182 |
0.023 |
| 302 |
250597_at |
AT5G07680
|
ANAC079, Arabidopsis NAC domain containing protein 79, ANAC080, NAC domain containing protein 80, ATNAC4, NAC080, NAC domain containing protein 80, NAC4 |
-0.179 |
0.089 |
| 303 |
258468_at |
AT3G06070
|
unknown |
-0.178 |
0.155 |
| 304 |
252501_at |
AT3G46880
|
unknown |
-0.178 |
0.015 |
| 305 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.176 |
-0.056 |
| 306 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.176 |
-0.330 |
| 307 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.176 |
-0.253 |
| 308 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
-0.175 |
-0.246 |
| 309 |
262399_at |
AT1G49500
|
unknown |
-0.174 |
-0.063 |
| 310 |
253515_at |
AT4G31320
|
SAUR37, SMALL AUXIN UPREGULATED RNA 37 |
-0.174 |
-0.004 |
| 311 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.173 |
-0.153 |
| 312 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
-0.173 |
-0.108 |
| 313 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
-0.172 |
-0.019 |
| 314 |
253036_at |
AT4G38340
|
NLP3, NIN-like protein 3 |
-0.172 |
0.002 |
| 315 |
259549_at |
AT1G35290
|
ALT1, acyl-lipid thioesterase 1 |
-0.172 |
-0.120 |
| 316 |
246844_at |
AT5G26660
|
ATMYB86, myb domain protein 86, MYB86, myb domain protein 86 |
-0.171 |
0.045 |
| 317 |
266259_at |
AT2G27830
|
unknown |
-0.170 |
0.125 |
| 318 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.169 |
-0.021 |
| 319 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.168 |
0.025 |
| 320 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
-0.167 |
0.011 |
| 321 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
-0.165 |
0.049 |
| 322 |
252180_at |
AT3G50630
|
ICK2, KRP2, KIP-related protein 2 |
-0.164 |
-0.044 |
| 323 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-0.164 |
-0.166 |
| 324 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.164 |
-0.206 |
| 325 |
257994_at |
AT3G19920
|
unknown |
-0.163 |
-0.049 |
| 326 |
249721_at |
AT5G24655
|
LSU4, RESPONSE TO LOW SULFUR 4 |
-0.163 |
-0.107 |
| 327 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.163 |
-0.119 |
| 328 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
-0.162 |
0.017 |
| 329 |
246439_at |
AT5G17600
|
[RING/U-box superfamily protein] |
-0.160 |
-0.172 |
| 330 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
-0.159 |
-0.110 |
| 331 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
-0.159 |
-0.366 |
| 332 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
-0.159 |
-0.051 |
| 333 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.157 |
0.203 |
| 334 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
-0.157 |
0.296 |