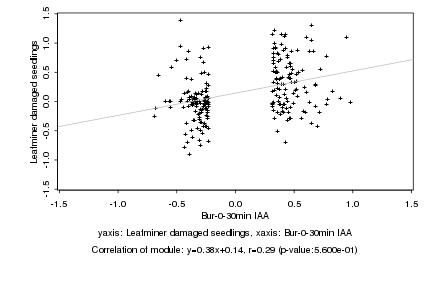
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Bur-0-30min IAA |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
0.978 |
-0.007 |
| 2 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
0.944 |
1.097 |
| 3 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.887 |
0.062 |
| 4 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.819 |
0.185 |
| 5 |
266611_at |
AT2G14960
|
GH3.1 |
0.779 |
0.043 |
| 6 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
0.772 |
-0.037 |
| 7 |
245076_at |
AT2G23170
|
GH3.3 |
0.769 |
0.781 |
| 8 |
253060_at |
AT4G37710
|
[VQ motif-containing protein] |
0.724 |
0.559 |
| 9 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
0.712 |
-0.174 |
| 10 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
0.695 |
-0.416 |
| 11 |
256009_at |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
0.681 |
0.304 |
| 12 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.678 |
0.280 |
| 13 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
0.676 |
-0.079 |
| 14 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.660 |
0.866 |
| 15 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.647 |
1.304 |
| 16 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
0.645 |
-0.363 |
| 17 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.643 |
1.058 |
| 18 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.629 |
0.864 |
| 19 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
0.624 |
-0.014 |
| 20 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.601 |
0.162 |
| 21 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.597 |
1.110 |
| 22 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
0.591 |
-0.180 |
| 23 |
265204_at |
AT2G36650
|
unknown |
0.585 |
0.252 |
| 24 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
0.575 |
-0.156 |
| 25 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.569 |
0.545 |
| 26 |
251178_at |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
0.557 |
-0.279 |
| 27 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.534 |
0.511 |
| 28 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.528 |
0.876 |
| 29 |
261476_at |
AT1G14480
|
[Ankyrin repeat family protein] |
0.524 |
0.097 |
| 30 |
262211_at |
AT1G74930
|
ORA47 |
0.518 |
0.467 |
| 31 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.517 |
0.344 |
| 32 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.512 |
0.207 |
| 33 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
0.511 |
0.189 |
| 34 |
250443_at |
AT5G10520
|
RBK1, ROP binding protein kinases 1 |
0.496 |
0.330 |
| 35 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.490 |
0.139 |
| 36 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
0.487 |
-0.031 |
| 37 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.482 |
0.559 |
| 38 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.476 |
0.858 |
| 39 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.474 |
0.489 |
| 40 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
0.470 |
0.333 |
| 41 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.467 |
0.651 |
| 42 |
255937_at |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
0.465 |
0.601 |
| 43 |
248801_at |
AT5G47370
|
HAT2 |
0.464 |
-0.278 |
| 44 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.464 |
0.401 |
| 45 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.461 |
-0.097 |
| 46 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.460 |
0.655 |
| 47 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
0.458 |
-0.289 |
| 48 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
0.454 |
0.399 |
| 49 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.453 |
0.468 |
| 50 |
252765_at |
AT3G42800
|
unknown |
0.449 |
-0.176 |
| 51 |
262085_at |
AT1G56060
|
unknown |
0.447 |
0.414 |
| 52 |
256442_at |
AT3G10930
|
unknown |
0.441 |
0.757 |
| 53 |
247869_at |
AT5G57520
|
ATZFP2, ZINC FINGER PROTEIN 2, ZFP2, zinc finger protein 2 |
0.439 |
0.005 |
| 54 |
253643_at |
AT4G29780
|
unknown |
0.438 |
0.791 |
| 55 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
0.436 |
-0.324 |
| 56 |
248789_at |
AT5G47440
|
unknown |
0.435 |
-0.116 |
| 57 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.432 |
0.063 |
| 58 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.431 |
0.597 |
| 59 |
253827_at |
AT4G28085
|
unknown |
0.425 |
0.924 |
| 60 |
263182_at |
AT1G05575
|
unknown |
0.421 |
1.162 |
| 61 |
253632_at |
AT4G30430
|
TET9, tetraspanin9 |
0.419 |
0.217 |
| 62 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
0.418 |
-0.689 |
| 63 |
248278_at |
AT5G52890
|
[AT hook motif-containing protein] |
0.411 |
-0.048 |
| 64 |
253832_at |
AT4G27654
|
unknown |
0.411 |
1.119 |
| 65 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.410 |
0.128 |
| 66 |
251246_at |
AT3G62100
|
IAA30, indole-3-acetic acid inducible 30 |
0.409 |
-0.036 |
| 67 |
254408_at |
AT4G21390
|
B120 |
0.408 |
0.882 |
| 68 |
265144_at |
AT1G51170
|
AGC2-3, AGC2 kinase 3, UCN, UNICORN |
0.407 |
-0.179 |
| 69 |
255844_at |
AT2G33580
|
LYK5, LysM-containing receptor-like kinase 5 |
0.402 |
0.304 |
| 70 |
258907_at |
AT3G06370
|
ATNHX4, NHX4, sodium hydrogen exchanger 4 |
0.397 |
-0.159 |
| 71 |
266507_at |
AT2G47860
|
SETH6 |
0.396 |
-0.091 |
| 72 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.395 |
0.417 |
| 73 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.392 |
1.153 |
| 74 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
0.390 |
0.307 |
| 75 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.389 |
0.981 |
| 76 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.382 |
0.406 |
| 77 |
260883_at |
AT1G29270
|
unknown |
0.380 |
-0.047 |
| 78 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.379 |
-0.208 |
| 79 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.379 |
0.728 |
| 80 |
251940_at |
AT3G53450
|
LOG4, LONELY GUY 4 |
0.375 |
0.078 |
| 81 |
261910_at |
AT1G80730
|
ATZFP1, ARABIDOPSIS THALIANA ZINC-FINGER PROTEIN 1, ZFP1, zinc-finger protein 1 |
0.372 |
-0.045 |
| 82 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.370 |
0.213 |
| 83 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.369 |
0.385 |
| 84 |
251488_at |
AT3G59440
|
[Calcium-binding EF-hand family protein] |
0.361 |
0.007 |
| 85 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.359 |
0.820 |
| 86 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
0.359 |
0.686 |
| 87 |
245755_at |
AT1G35210
|
unknown |
0.358 |
0.313 |
| 88 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.358 |
0.826 |
| 89 |
247913_at |
AT5G57510
|
unknown |
0.356 |
0.239 |
| 90 |
253791_at |
AT4G28640
|
IAA11, indole-3-acetic acid inducible 11 |
0.352 |
-0.178 |
| 91 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
0.352 |
-0.500 |
| 92 |
253044_at |
AT4G37290
|
unknown |
0.351 |
0.512 |
| 93 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
0.349 |
0.387 |
| 94 |
258787_at |
AT3G11840
|
PUB24, plant U-box 24 |
0.346 |
0.519 |
| 95 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.345 |
0.397 |
| 96 |
265327_at |
AT2G18210
|
unknown |
0.342 |
0.117 |
| 97 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
0.342 |
0.500 |
| 98 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.339 |
0.920 |
| 99 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.339 |
0.507 |
| 100 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.339 |
0.596 |
| 101 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.333 |
0.939 |
| 102 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
0.331 |
0.191 |
| 103 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.331 |
1.221 |
| 104 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
0.330 |
-0.276 |
| 105 |
247047_at |
AT5G66650
|
unknown |
0.325 |
0.999 |
| 106 |
267623_at |
AT2G39650
|
unknown |
0.324 |
0.339 |
| 107 |
252053_at |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
0.324 |
0.653 |
| 108 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.322 |
0.915 |
| 109 |
251603_at |
AT3G57760
|
[Protein kinase superfamily protein] |
0.321 |
0.517 |
| 110 |
250796_at |
AT5G05300
|
unknown |
0.321 |
0.524 |
| 111 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.320 |
-0.137 |
| 112 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
0.320 |
-0.009 |
| 113 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.318 |
0.716 |
| 114 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.317 |
0.828 |
| 115 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
0.316 |
0.767 |
| 116 |
267069_at |
AT2G41010
|
ATCAMBP25, calmodulin (CAM)-binding protein of 25 kDa, CAMBP25, calmodulin (CAM)-binding protein of 25 kDa |
0.314 |
-0.024 |
| 117 |
256010_at |
AT1G19220
|
ARF11, AUXIN RESPONSE FACTOR11, ARF19, auxin response factor 19, IAA22, indole-3-acetic acid inducible 22 |
0.314 |
-0.040 |
| 118 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.312 |
0.181 |
| 119 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.311 |
1.160 |
| 120 |
266908_at |
AT2G34650
|
ABR, ABRUPTUS, PID, PINOID |
0.310 |
-0.080 |
| 121 |
256190_at |
AT1G30100
|
ATNCED5, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 5, NCED5, nine-cis-epoxycarotenoid dioxygenase 5 |
0.308 |
-0.024 |
| 122 |
265387_at |
AT2G20670
|
unknown |
-0.691 |
-0.242 |
| 123 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.683 |
-0.105 |
| 124 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.660 |
0.446 |
| 125 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.600 |
0.011 |
| 126 |
261567_at |
AT1G33055
|
unknown |
-0.566 |
0.015 |
| 127 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
-0.560 |
-0.091 |
| 128 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
-0.556 |
0.004 |
| 129 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.552 |
0.595 |
| 130 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
-0.508 |
0.709 |
| 131 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
-0.476 |
1.403 |
| 132 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.473 |
0.013 |
| 133 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.469 |
0.951 |
| 134 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.465 |
0.039 |
| 135 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.443 |
-0.088 |
| 136 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.441 |
-0.783 |
| 137 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
-0.438 |
0.147 |
| 138 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.433 |
-0.561 |
| 139 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.422 |
0.732 |
| 140 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.421 |
0.405 |
| 141 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
-0.420 |
-0.376 |
| 142 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.413 |
-0.688 |
| 143 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.411 |
0.005 |
| 144 |
258225_at |
AT3G15630
|
unknown |
-0.410 |
0.156 |
| 145 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
-0.407 |
0.861 |
| 146 |
254954_at |
AT4G10910
|
unknown |
-0.405 |
-0.076 |
| 147 |
247754_at |
AT5G59080
|
unknown |
-0.404 |
0.178 |
| 148 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.398 |
0.073 |
| 149 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.396 |
-0.892 |
| 150 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
-0.393 |
0.043 |
| 151 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
-0.393 |
0.069 |
| 152 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.381 |
0.384 |
| 153 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.380 |
-0.050 |
| 154 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.379 |
-0.492 |
| 155 |
260081_at |
AT1G78170
|
unknown |
-0.376 |
0.087 |
| 156 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.373 |
-0.012 |
| 157 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-0.370 |
-0.056 |
| 158 |
263208_at |
AT1G10480
|
ZFP5, zinc finger protein 5 |
-0.368 |
0.016 |
| 159 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.366 |
-0.610 |
| 160 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
-0.365 |
-0.027 |
| 161 |
251434_at |
AT3G59850
|
[Pectin lyase-like superfamily protein] |
-0.363 |
0.053 |
| 162 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.360 |
-0.309 |
| 163 |
253155_at |
AT4G35720
|
unknown |
-0.356 |
0.075 |
| 164 |
249768_at |
AT5G24100
|
[Leucine-rich repeat protein kinase family protein] |
-0.352 |
0.036 |
| 165 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-0.350 |
-0.319 |
| 166 |
267228_at |
AT2G43890
|
[Pectin lyase-like superfamily protein] |
-0.347 |
0.017 |
| 167 |
253305_at |
AT4G33666
|
unknown |
-0.347 |
-0.156 |
| 168 |
265539_at |
AT2G15830
|
unknown |
-0.342 |
0.140 |
| 169 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.340 |
0.066 |
| 170 |
247882_at |
AT5G57785
|
unknown |
-0.340 |
-0.084 |
| 171 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.337 |
0.123 |
| 172 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.335 |
-0.048 |
| 173 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.332 |
-0.022 |
| 174 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.331 |
0.015 |
| 175 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
-0.327 |
-0.459 |
| 176 |
253814_at |
AT4G28290
|
unknown |
-0.318 |
0.149 |
| 177 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
-0.318 |
-0.216 |
| 178 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.314 |
-0.305 |
| 179 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
-0.314 |
0.003 |
| 180 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.311 |
-0.272 |
| 181 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.310 |
-0.192 |
| 182 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.309 |
-0.650 |
| 183 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
-0.302 |
0.767 |
| 184 |
263970_at |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
-0.301 |
-0.029 |
| 185 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.300 |
-0.748 |
| 186 |
265584_at |
AT2G20180
|
PIF1, PHY-INTERACTING FACTOR 1, PIL5, phytochrome interacting factor 3-like 5 |
-0.299 |
-0.488 |
| 187 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.298 |
-0.133 |
| 188 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.298 |
-0.343 |
| 189 |
249493_at |
AT5G39080
|
[HXXXD-type acyl-transferase family protein] |
-0.296 |
-0.353 |
| 190 |
246142_at |
AT5G19970
|
unknown |
-0.295 |
-0.183 |
| 191 |
262170_at |
AT1G74940
|
unknown |
-0.295 |
-0.139 |
| 192 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.293 |
0.487 |
| 193 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
-0.290 |
0.134 |
| 194 |
264463_at |
AT1G10150
|
[Carbohydrate-binding protein] |
-0.285 |
-0.134 |
| 195 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.285 |
0.172 |
| 196 |
264909_at |
AT2G17300
|
unknown |
-0.282 |
-0.537 |
| 197 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.280 |
-0.423 |
| 198 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
-0.279 |
0.675 |
| 199 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
-0.279 |
-0.081 |
| 200 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.273 |
0.916 |
| 201 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
-0.272 |
-0.048 |
| 202 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
-0.272 |
-0.020 |
| 203 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.272 |
-0.365 |
| 204 |
254743_at |
AT4G13420
|
ATHAK5, ARABIDOPSIS THALIANA HIGH AFFINITY K+ TRANSPORTER 5, HAK5, high affinity K+ transporter 5 |
-0.266 |
0.007 |
| 205 |
245412_at |
AT4G17280
|
[Auxin-responsive family protein] |
-0.266 |
0.027 |
| 206 |
265481_at |
AT2G15960
|
unknown |
-0.264 |
0.500 |
| 207 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
-0.261 |
0.177 |
| 208 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.261 |
-0.050 |
| 209 |
247585_at |
AT5G60680
|
unknown |
-0.259 |
0.060 |
| 210 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.259 |
-0.425 |
| 211 |
252057_at |
AT3G52480
|
unknown |
-0.258 |
-0.112 |
| 212 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.257 |
0.081 |
| 213 |
249377_at |
AT5G40690
|
unknown |
-0.256 |
-0.049 |
| 214 |
265031_at |
AT1G61590
|
[Protein kinase superfamily protein] |
-0.255 |
-0.018 |
| 215 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.255 |
-0.023 |
| 216 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.253 |
0.315 |
| 217 |
250597_at |
AT5G07680
|
ANAC079, Arabidopsis NAC domain containing protein 79, ANAC080, NAC domain containing protein 80, ATNAC4, NAC080, NAC domain containing protein 80, NAC4 |
-0.252 |
0.071 |
| 218 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
-0.251 |
-0.006 |
| 219 |
262399_at |
AT1G49500
|
unknown |
-0.251 |
0.187 |
| 220 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
-0.250 |
-0.118 |
| 221 |
258038_at |
AT3G21260
|
GLTP3, GLYCOLIPID TRANSFER PROTEIN 3 |
-0.249 |
-0.235 |
| 222 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
-0.249 |
0.223 |
| 223 |
253839_at |
AT4G27890
|
[HSP20-like chaperones superfamily protein] |
-0.247 |
-0.015 |
| 224 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.247 |
-0.416 |
| 225 |
252036_at |
AT3G52070
|
unknown |
-0.245 |
0.095 |
| 226 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.244 |
-0.143 |
| 227 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.244 |
-0.137 |
| 228 |
255538_at |
AT4G01680
|
AtMYB55, myb domain protein 55, MYB55, myb domain protein 55 |
-0.243 |
-0.277 |
| 229 |
259017_at |
AT3G07310
|
unknown |
-0.243 |
-0.179 |
| 230 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
-0.243 |
-0.029 |
| 231 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.242 |
-0.256 |
| 232 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.241 |
0.005 |
| 233 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
-0.238 |
0.928 |
| 234 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.238 |
-0.455 |
| 235 |
253812_at |
AT4G28240
|
[Wound-responsive family protein] |
-0.238 |
-0.001 |
| 236 |
264210_at |
AT1G22640
|
ATMYB3, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 3, MYB3, myb domain protein 3 |
-0.237 |
0.276 |
| 237 |
252659_at |
AT3G44430
|
unknown |
-0.236 |
-0.076 |
| 238 |
252232_at |
AT3G49760
|
AtbZIP5, basic leucine-zipper 5, bZIP5, basic leucine-zipper 5 |
-0.235 |
0.059 |
| 239 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.234 |
-0.035 |
| 240 |
249941_at |
AT5G22270
|
unknown |
-0.234 |
0.470 |
| 241 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.234 |
-0.673 |