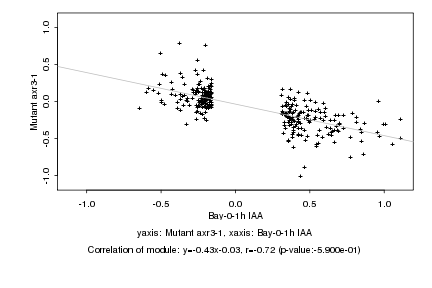
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Bay-0-1h IAA |
Log2 signal ratio
Mutant axr3-1 |
| 1 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
1.107 |
-0.230 |
| 2 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
1.106 |
-0.495 |
| 3 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
1.053 |
-0.576 |
| 4 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
1.006 |
-0.306 |
| 5 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
0.992 |
-0.300 |
| 6 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
0.968 |
-0.462 |
| 7 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.959 |
0.009 |
| 8 |
247869_at |
AT5G57520
|
ATZFP2, ZINC FINGER PROTEIN 2, ZFP2, zinc finger protein 2 |
0.954 |
-0.412 |
| 9 |
254459_at |
AT4G21200
|
ATGA2OX8, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 8, GA2OX8, gibberellin 2-oxidase 8 |
0.864 |
-0.295 |
| 10 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.860 |
-0.713 |
| 11 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.845 |
-0.528 |
| 12 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
0.841 |
-0.411 |
| 13 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
0.839 |
-0.370 |
| 14 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
0.809 |
-0.277 |
| 15 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
0.808 |
-0.205 |
| 16 |
245076_at |
AT2G23170
|
GH3.3 |
0.783 |
-0.159 |
| 17 |
266611_at |
AT2G14960
|
GH3.1 |
0.773 |
-0.476 |
| 18 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
0.767 |
-0.750 |
| 19 |
251178_at |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
0.720 |
-0.185 |
| 20 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
0.720 |
-0.356 |
| 21 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.697 |
-0.296 |
| 22 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.696 |
-0.308 |
| 23 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
0.691 |
-0.188 |
| 24 |
252103_at |
AT3G51410
|
unknown |
0.687 |
-0.403 |
| 25 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
0.683 |
-0.383 |
| 26 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
0.676 |
-0.337 |
| 27 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
0.668 |
-0.181 |
| 28 |
247474_at |
AT5G62280
|
unknown |
0.665 |
-0.247 |
| 29 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
0.664 |
-0.541 |
| 30 |
259785_at |
AT1G29490
|
SAUR68, SMALL AUXIN UPREGULATED 68 |
0.661 |
-0.382 |
| 31 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
0.649 |
-0.397 |
| 32 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.640 |
-0.406 |
| 33 |
253179_at |
AT4G35200
|
unknown |
0.640 |
-0.458 |
| 34 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.633 |
-0.358 |
| 35 |
259027_at |
AT3G09280
|
unknown |
0.633 |
-0.251 |
| 36 |
246798_at |
AT5G26930
|
GATA23, GATA transcription factor 23 |
0.620 |
-0.432 |
| 37 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
0.611 |
-0.342 |
| 38 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
0.605 |
-0.088 |
| 39 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
0.599 |
-0.189 |
| 40 |
256190_at |
AT1G30100
|
ATNCED5, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 5, NCED5, nine-cis-epoxycarotenoid dioxygenase 5 |
0.597 |
-0.136 |
| 41 |
248278_at |
AT5G52890
|
[AT hook motif-containing protein] |
0.589 |
-0.021 |
| 42 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
0.588 |
-0.247 |
| 43 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.585 |
-0.485 |
| 44 |
253180_at |
AT4G35210
|
unknown |
0.568 |
-0.148 |
| 45 |
248801_at |
AT5G47370
|
HAT2 |
0.567 |
-0.225 |
| 46 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
0.562 |
-0.383 |
| 47 |
252765_at |
AT3G42800
|
unknown |
0.554 |
-0.554 |
| 48 |
262445_at |
AT1G47485
|
unknown |
0.552 |
-0.105 |
| 49 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.545 |
-0.455 |
| 50 |
247878_at |
AT5G57760
|
unknown |
0.542 |
-0.577 |
| 51 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
0.540 |
-0.600 |
| 52 |
246992_at |
AT5G67430
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.537 |
-0.133 |
| 53 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.535 |
-0.006 |
| 54 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
0.535 |
-0.204 |
| 55 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.529 |
-0.236 |
| 56 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.506 |
-0.112 |
| 57 |
248789_at |
AT5G47440
|
unknown |
0.503 |
0.023 |
| 58 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
0.499 |
-0.250 |
| 59 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.495 |
-0.277 |
| 60 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
0.490 |
-0.176 |
| 61 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
0.486 |
-0.113 |
| 62 |
245075_at |
AT2G23180
|
CYP96A1, cytochrome P450, family 96, subfamily A, polypeptide 1 |
0.485 |
-0.243 |
| 63 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.482 |
-0.187 |
| 64 |
250327_at |
AT5G12050
|
unknown |
0.482 |
-0.459 |
| 65 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.478 |
0.114 |
| 66 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
0.473 |
-0.056 |
| 67 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.472 |
-0.219 |
| 68 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
0.470 |
-0.525 |
| 69 |
245108_at |
AT2G41510
|
ATCKX1, CYTOKININ OXIDASE/DEHYDROGENASE 1, CKX1, cytokinin oxidase/dehydrogenase 1 |
0.464 |
-0.218 |
| 70 |
253413_at |
AT4G33020
|
ATZIP9, ZIP9 |
0.464 |
0.019 |
| 71 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.459 |
-0.379 |
| 72 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
0.458 |
-0.879 |
| 73 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
0.457 |
-0.142 |
| 74 |
255028_at |
AT4G09890
|
unknown |
0.441 |
-0.354 |
| 75 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
0.440 |
-0.143 |
| 76 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
0.438 |
-0.454 |
| 77 |
246652_at |
AT5G35190
|
EXT13, extensin 13 |
0.435 |
-1.001 |
| 78 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
0.434 |
-0.147 |
| 79 |
251246_at |
AT3G62100
|
IAA30, indole-3-acetic acid inducible 30 |
0.434 |
-0.295 |
| 80 |
266507_at |
AT2G47860
|
SETH6 |
0.425 |
-0.156 |
| 81 |
257386_at |
AT2G42440
|
ASL15, ASYMMETRIC LEAVES 2-like 15, LBD17, LOB domain-containing protein 17 |
0.422 |
-0.276 |
| 82 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
0.422 |
-0.364 |
| 83 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.421 |
-0.180 |
| 84 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.420 |
0.130 |
| 85 |
265144_at |
AT1G51170
|
AGC2-3, AGC2 kinase 3, UCN, UNICORN |
0.420 |
-0.231 |
| 86 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
0.420 |
-0.437 |
| 87 |
263576_at |
AT2G17080
|
unknown |
0.419 |
-0.260 |
| 88 |
248509_at |
AT5G50335
|
unknown |
0.419 |
-0.452 |
| 89 |
260058_at |
AT1G78100
|
AUF1, auxin up-regulated f-box protein 1 |
0.417 |
-0.190 |
| 90 |
256010_at |
AT1G19220
|
ARF11, AUXIN RESPONSE FACTOR11, ARF19, auxin response factor 19, IAA22, indole-3-acetic acid inducible 22 |
0.411 |
-0.204 |
| 91 |
247622_at |
AT5G60350
|
unknown |
0.407 |
-0.049 |
| 92 |
249109_at |
AT5G43700
|
ATAUX2-11, AUXIN INDUCIBLE 2-11, IAA4, indole-3-acetic acid inducible 4 |
0.406 |
-0.309 |
| 93 |
258253_at |
AT3G26760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.401 |
-0.324 |
| 94 |
253791_at |
AT4G28640
|
IAA11, indole-3-acetic acid inducible 11 |
0.398 |
-0.112 |
| 95 |
256872_at |
AT3G26490
|
[Phototropic-responsive NPH3 family protein] |
0.396 |
0.027 |
| 96 |
258907_at |
AT3G06370
|
ATNHX4, NHX4, sodium hydrogen exchanger 4 |
0.396 |
-0.264 |
| 97 |
248317_at |
AT5G52680
|
[Copper transport protein family] |
0.394 |
0.047 |
| 98 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.390 |
-0.350 |
| 99 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
0.389 |
-0.414 |
| 100 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
0.386 |
-0.614 |
| 101 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
0.386 |
-0.386 |
| 102 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.385 |
-0.254 |
| 103 |
257026_at |
AT3G19200
|
unknown |
0.384 |
-0.135 |
| 104 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
0.383 |
-0.235 |
| 105 |
261466_at |
AT1G07690
|
unknown |
0.381 |
-0.039 |
| 106 |
262045_at |
AT1G80240
|
DGR1, DUF642 L-GalL responsive gene 1 |
0.381 |
-0.477 |
| 107 |
257975_at |
AT3G20830
|
AGC2-4, AGC2 kinase 4, UCNL, UNICORN-like |
0.380 |
-0.210 |
| 108 |
256799_at |
AT3G18560
|
unknown |
0.377 |
-0.437 |
| 109 |
265447_at |
AT2G46570
|
LAC6, laccase 6 |
0.377 |
-0.149 |
| 110 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
0.373 |
-0.301 |
| 111 |
261910_at |
AT1G80730
|
ATZFP1, ARABIDOPSIS THALIANA ZINC-FINGER PROTEIN 1, ZFP1, zinc-finger protein 1 |
0.373 |
-0.057 |
| 112 |
245416_at |
AT4G17350
|
unknown |
0.371 |
-0.273 |
| 113 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
0.369 |
0.168 |
| 114 |
252557_at |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
0.368 |
0.037 |
| 115 |
251940_at |
AT3G53450
|
LOG4, LONELY GUY 4 |
0.368 |
-0.105 |
| 116 |
252279_at |
AT3G49700
|
ACS9, 1-aminocyclopropane-1-carboxylate synthase 9, AtACS9, ETO3, ETHYLENE OVERPRODUCING 3 |
0.367 |
-0.217 |
| 117 |
251925_at |
AT3G54000
|
unknown |
0.363 |
-0.143 |
| 118 |
248423_at |
AT5G51670
|
unknown |
0.362 |
-0.319 |
| 119 |
259735_at |
AT1G64405
|
unknown |
0.361 |
-0.249 |
| 120 |
246534_at |
AT5G15890
|
TBL21, TRICHOME BIREFRINGENCE-LIKE 21 |
0.360 |
-0.127 |
| 121 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
0.360 |
-0.090 |
| 122 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
0.358 |
-0.228 |
| 123 |
258388_at |
AT3G15370
|
ATEXP12, ATEXPA12, expansin 12, ATHEXP ALPHA 1.24, EXP12, EXPANSIN 12, EXPA12, expansin 12 |
0.356 |
-0.193 |
| 124 |
251751_at |
AT3G55720
|
unknown |
0.355 |
-0.281 |
| 125 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
0.355 |
-0.514 |
| 126 |
252364_at |
AT3G48450
|
[RPM1-interacting protein 4 (RIN4) family protein] |
0.355 |
0.056 |
| 127 |
265272_at |
AT2G28350
|
ARF10, auxin response factor 10 |
0.351 |
-0.035 |
| 128 |
251423_at |
AT3G60550
|
CYCP3;2, cyclin p3;2 |
0.350 |
-0.527 |
| 129 |
251160_at |
AT3G63240
|
[DNAse I-like superfamily protein] |
0.345 |
-0.204 |
| 130 |
253011_at |
AT4G37890
|
EDA40, embryo sac development arrest 40 |
0.343 |
-0.192 |
| 131 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.342 |
-0.150 |
| 132 |
266908_at |
AT2G34650
|
ABR, ABRUPTUS, PID, PINOID |
0.337 |
-0.066 |
| 133 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.337 |
-0.008 |
| 134 |
252204_at |
AT3G50340
|
unknown |
0.336 |
-0.167 |
| 135 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
0.335 |
-0.160 |
| 136 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
0.333 |
-0.364 |
| 137 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
0.331 |
-0.049 |
| 138 |
255788_at |
AT2G33310
|
IAA13, auxin-induced protein 13 |
0.327 |
-0.274 |
| 139 |
250723_at |
AT5G06300
|
LOG7, LONELY GUY 7 |
0.326 |
-0.323 |
| 140 |
259845_at |
AT1G73590
|
ATPIN1, ARABIDOPSIS THALIANA PIN-FORMED 1, PIN1, PIN-FORMED 1 |
0.325 |
-0.171 |
| 141 |
258782_at |
AT3G11750
|
FOLB1 |
0.322 |
-0.154 |
| 142 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
0.320 |
-0.429 |
| 143 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
0.316 |
-0.168 |
| 144 |
262263_at |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
0.315 |
-0.131 |
| 145 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
0.314 |
-0.064 |
| 146 |
266625_at |
AT2G35380
|
[Peroxidase superfamily protein] |
0.313 |
0.164 |
| 147 |
260494_at |
AT2G41820
|
PXC3, PXY/TDR-correlated 3 |
0.307 |
-0.152 |
| 148 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.303 |
0.088 |
| 149 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.650 |
-0.083 |
| 150 |
261567_at |
AT1G33055
|
unknown |
-0.602 |
0.134 |
| 151 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.585 |
0.181 |
| 152 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.557 |
0.152 |
| 153 |
247070_at |
AT5G66815
|
unknown |
-0.523 |
0.109 |
| 154 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.513 |
0.242 |
| 155 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
-0.504 |
0.652 |
| 156 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
-0.499 |
-0.009 |
| 157 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
-0.499 |
0.024 |
| 158 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
-0.493 |
0.377 |
| 159 |
256631_at |
AT3G28320
|
unknown |
-0.482 |
-0.034 |
| 160 |
254200_at |
AT4G24110
|
unknown |
-0.475 |
0.352 |
| 161 |
250472_at |
AT5G10210
|
unknown |
-0.435 |
0.095 |
| 162 |
247024_at |
AT5G66985
|
unknown |
-0.432 |
0.270 |
| 163 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
-0.428 |
0.171 |
| 164 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.405 |
0.082 |
| 165 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-0.392 |
-0.093 |
| 166 |
253060_at |
AT4G37710
|
[VQ motif-containing protein] |
-0.392 |
-0.007 |
| 167 |
263829_at |
AT2G40435
|
unknown |
-0.381 |
0.786 |
| 168 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
-0.376 |
0.384 |
| 169 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.374 |
0.033 |
| 170 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.371 |
-0.116 |
| 171 |
267228_at |
AT2G43890
|
[Pectin lyase-like superfamily protein] |
-0.371 |
0.107 |
| 172 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.361 |
0.075 |
| 173 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
-0.360 |
0.326 |
| 174 |
251434_at |
AT3G59850
|
[Pectin lyase-like superfamily protein] |
-0.351 |
-0.044 |
| 175 |
250582_at |
AT5G07580
|
[Integrase-type DNA-binding superfamily protein] |
-0.347 |
0.091 |
| 176 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.347 |
0.242 |
| 177 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
-0.346 |
0.087 |
| 178 |
245258_at |
AT4G15340
|
04C11, ATPEN1, pentacyclic triterpene synthase 1, PEN1, pentacyclic triterpene synthase 1 |
-0.335 |
-0.306 |
| 179 |
245169_at |
AT2G33220
|
[GRIM-19 protein] |
-0.333 |
0.014 |
| 180 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
-0.328 |
0.054 |
| 181 |
259520_at |
AT1G12320
|
unknown |
-0.326 |
0.003 |
| 182 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
-0.311 |
-0.041 |
| 183 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.292 |
0.050 |
| 184 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
-0.289 |
0.167 |
| 185 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
-0.287 |
0.131 |
| 186 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
-0.286 |
0.152 |
| 187 |
247213_at |
AT5G64900
|
ATPEP1, ARABIDOPSIS THALIANA PEPTIDE 1, PEP1, PEPTIDE 1, PROPEP1, precursor of peptide 1 |
-0.274 |
0.103 |
| 188 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
-0.272 |
0.126 |
| 189 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
-0.269 |
0.421 |
| 190 |
247991_at |
AT5G56320
|
ATEXP14, ATEXPA14, expansin A14, ATHEXP ALPHA 1.5, EXP14, EXPANSIN 14, EXPA14, expansin A14 |
-0.267 |
-0.241 |
| 191 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
-0.266 |
-0.141 |
| 192 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
-0.266 |
-0.047 |
| 193 |
256358_at |
AT1G66470
|
AtRHD6, RHD6, ROOT HAIR DEFECTIVE6 |
-0.264 |
0.130 |
| 194 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.264 |
-0.144 |
| 195 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
-0.263 |
0.149 |
| 196 |
252574_at |
AT3G45430
|
LecRK-I.5, L-type lectin receptor kinase I.5 |
-0.261 |
-0.124 |
| 197 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
-0.261 |
0.554 |
| 198 |
256788_at |
AT3G13730
|
CYP90D1, cytochrome P450, family 90, subfamily D, polypeptide 1 |
-0.258 |
0.182 |
| 199 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
-0.257 |
-0.069 |
| 200 |
263380_at |
AT2G40200
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.256 |
0.377 |
| 201 |
265615_at |
AT2G25450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.253 |
-0.052 |
| 202 |
254175_at |
AT4G24050
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.253 |
0.125 |
| 203 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
-0.253 |
0.235 |
| 204 |
267216_at |
AT2G02620
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.252 |
0.015 |
| 205 |
247827_at |
AT5G58500
|
LSH5, LIGHT SENSITIVE HYPOCOTYLS 5 |
-0.252 |
0.144 |
| 206 |
259371_at |
AT1G69080
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.247 |
-0.054 |
| 207 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.242 |
-0.012 |
| 208 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
-0.242 |
0.230 |
| 209 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.241 |
-0.153 |
| 210 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
-0.237 |
0.272 |
| 211 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
-0.234 |
0.011 |
| 212 |
253400_at |
AT4G32860
|
unknown |
-0.234 |
0.053 |
| 213 |
251785_at |
AT3G55130
|
ABCG19, ATP-binding cassette G19, ATWBC19, white-brown complex homolog 19, WBC19, white-brown complex homolog 19 |
-0.234 |
0.176 |
| 214 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.232 |
-0.031 |
| 215 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.231 |
0.017 |
| 216 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
-0.229 |
0.047 |
| 217 |
262211_at |
AT1G74930
|
ORA47 |
-0.227 |
0.029 |
| 218 |
250205_at |
AT5G14020
|
[Endosomal targeting BRO1-like domain-containing protein] |
-0.226 |
-0.165 |
| 219 |
259982_at |
AT1G76410
|
ATL8 |
-0.225 |
0.086 |
| 220 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
-0.224 |
-0.069 |
| 221 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
-0.223 |
0.005 |
| 222 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
-0.223 |
-0.023 |
| 223 |
257925_at |
AT3G23170
|
unknown |
-0.222 |
0.133 |
| 224 |
252938_at |
AT4G39190
|
unknown |
-0.221 |
-0.025 |
| 225 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
-0.219 |
0.432 |
| 226 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
-0.218 |
-0.012 |
| 227 |
252180_at |
AT3G50630
|
ICK2, KRP2, KIP-related protein 2 |
-0.216 |
0.081 |
| 228 |
253017_at |
AT4G37970
|
ATCAD6, CAD6, cinnamyl alcohol dehydrogenase 6 |
-0.216 |
0.165 |
| 229 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.214 |
0.028 |
| 230 |
244921_s_at |
ATMG01000
|
ORF114 |
-0.214 |
-0.058 |
| 231 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.214 |
0.181 |
| 232 |
247492_at |
AT5G61890
|
[Integrase-type DNA-binding superfamily protein] |
-0.214 |
0.087 |
| 233 |
262170_at |
AT1G74940
|
unknown |
-0.213 |
0.157 |
| 234 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.212 |
-0.223 |
| 235 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.210 |
0.179 |
| 236 |
263176_at |
AT1G05530
|
UGT2, UDP-GLUCOSYL TRANSFERASE 2, UGT75B2, UDP-glucosyl transferase 75B2 |
-0.209 |
-0.007 |
| 237 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
-0.208 |
0.003 |
| 238 |
246580_at |
AT1G31770
|
ABCG14, ATP-binding cassette G14 |
-0.206 |
0.022 |
| 239 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
-0.206 |
0.761 |
| 240 |
260491_at |
AT1G51440
|
DALL2, DAD1-Like Lipase 2 |
-0.205 |
0.027 |
| 241 |
248408_at |
AT5G51520
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.205 |
0.079 |
| 242 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
-0.205 |
0.111 |
| 243 |
265305_at |
AT2G20340
|
AAS, aromatic aldehyde synthase, AtAAS |
-0.205 |
0.090 |
| 244 |
265057_at |
AT1G52140
|
unknown |
-0.204 |
0.052 |
| 245 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.204 |
-0.026 |
| 246 |
256382_at |
AT1G66860
|
[Class I glutamine amidotransferase-like superfamily protein] |
-0.203 |
-0.067 |
| 247 |
253830_at |
AT4G27652
|
unknown |
-0.203 |
0.135 |
| 248 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
-0.202 |
-0.138 |
| 249 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
-0.201 |
-0.248 |
| 250 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.201 |
0.076 |
| 251 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.200 |
0.123 |
| 252 |
263297_at |
AT2G15310
|
ARFB1A, ADP-ribosylation factor B1A, ATARFB1A, ADP-ribosylation factor B1A |
-0.198 |
0.112 |
| 253 |
265184_at |
AT1G23710
|
unknown |
-0.196 |
-0.074 |
| 254 |
266066_at |
AT2G18800
|
ATXTH21, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 21, XTH21, xyloglucan endotransglucosylase/hydrolase 21 |
-0.195 |
-0.090 |
| 255 |
253525_at |
AT4G31330
|
unknown |
-0.194 |
0.311 |
| 256 |
258442_at |
AT3G01015
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.194 |
-0.028 |
| 257 |
263182_at |
AT1G05575
|
unknown |
-0.193 |
0.033 |
| 258 |
254258_at |
AT4G23410
|
TET5, tetraspanin5 |
-0.192 |
0.127 |
| 259 |
265124_at |
AT1G55430
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.192 |
0.115 |
| 260 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
-0.191 |
0.096 |
| 261 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.188 |
-0.063 |
| 262 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
-0.188 |
0.086 |
| 263 |
254632_at |
AT4G18630
|
unknown |
-0.186 |
-0.091 |
| 264 |
253101_at |
AT4G37430
|
CYP81F1, CYTOCHROME P450 MONOOXYGENASE 81F1, CYP91A2, cytochrome P450, family 91, subfamily A, polypeptide 2 |
-0.184 |
0.141 |
| 265 |
250719_at |
AT5G06250
|
DPA4, DEVELOPMENT-RELATED PcG TARGET IN THE APEX 4 |
-0.183 |
0.088 |
| 266 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
-0.183 |
0.205 |
| 267 |
251116_at |
AT3G63470
|
scpl40, serine carboxypeptidase-like 40 |
-0.181 |
0.045 |
| 268 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
-0.181 |
0.182 |
| 269 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.180 |
0.092 |
| 270 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.178 |
0.055 |
| 271 |
266007_at |
AT2G37380
|
MAKR3, MEMBRANE-ASSOCIATED KINASE REGULATOR 3 |
-0.176 |
-0.052 |
| 272 |
266108_at |
AT2G37900
|
[Major facilitator superfamily protein] |
-0.176 |
-0.012 |
| 273 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.175 |
0.072 |
| 274 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
-0.175 |
0.085 |
| 275 |
263831_at |
AT2G40300
|
ATFER4, ferritin 4, FER4, ferritin 4 |
-0.175 |
0.073 |
| 276 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
-0.175 |
-0.058 |
| 277 |
246216_at |
AT4G36380
|
ROT3, ROTUNDIFOLIA 3 |
-0.173 |
0.170 |
| 278 |
245585_at |
AT4G14970
|
unknown |
-0.172 |
0.048 |
| 279 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
-0.171 |
0.095 |
| 280 |
261924_at |
AT1G22550
|
[Major facilitator superfamily protein] |
-0.171 |
0.050 |
| 281 |
263772_at |
AT2G06255
|
ELF4-L3, ELF4-like 3 |
-0.171 |
-0.058 |
| 282 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.169 |
0.021 |
| 283 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.169 |
0.131 |
| 284 |
257375_at |
AT2G38640
|
unknown |
-0.168 |
0.217 |
| 285 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
-0.168 |
0.020 |
| 286 |
259302_at |
AT3G05120
|
ATGID1A, GA INSENSITIVE DWARF1A, GID1A, GA INSENSITIVE DWARF1A |
-0.167 |
0.307 |
| 287 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
-0.167 |
0.121 |
| 288 |
247047_at |
AT5G66650
|
unknown |
-0.167 |
-0.012 |
| 289 |
249768_at |
AT5G24100
|
[Leucine-rich repeat protein kinase family protein] |
-0.166 |
0.065 |
| 290 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.166 |
-0.074 |
| 291 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.165 |
-0.051 |
| 292 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
-0.165 |
0.249 |
| 293 |
255942_at |
AT1G22360
|
AtUGT85A2, UDP-glucosyl transferase 85A2, UGT85A2, UDP-glucosyl transferase 85A2 |
-0.164 |
0.216 |
| 294 |
248140_at |
AT5G54980
|
[Uncharacterised protein family (UPF0497)] |
-0.164 |
-0.027 |
| 295 |
266954_at |
AT2G34530
|
unknown |
-0.164 |
0.122 |