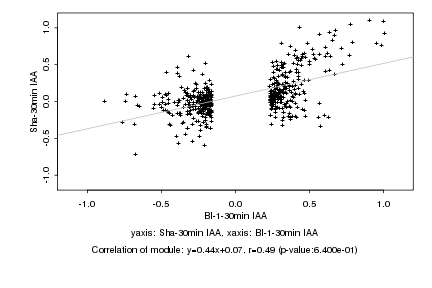
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
BI-1-30min IAA |
Log2 signal ratio
Sha-30min IAA |
| 1 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
1.003 |
0.936 |
| 2 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.994 |
1.096 |
| 3 |
259785_at |
AT1G29490
|
SAUR68, SMALL AUXIN UPREGULATED 68 |
0.982 |
0.763 |
| 4 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
0.951 |
0.794 |
| 5 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
0.902 |
1.113 |
| 6 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
0.786 |
0.804 |
| 7 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
0.771 |
1.051 |
| 8 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
0.765 |
0.625 |
| 9 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.718 |
0.510 |
| 10 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
0.716 |
0.724 |
| 11 |
245076_at |
AT2G23170
|
GH3.3 |
0.670 |
0.969 |
| 12 |
266611_at |
AT2G14960
|
GH3.1 |
0.666 |
0.370 |
| 13 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
0.665 |
0.900 |
| 14 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
0.653 |
0.831 |
| 15 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
0.640 |
0.623 |
| 16 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.635 |
0.943 |
| 17 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
0.631 |
0.433 |
| 18 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
0.625 |
-0.211 |
| 19 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
0.620 |
0.654 |
| 20 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.606 |
0.415 |
| 21 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.604 |
0.618 |
| 22 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.604 |
0.741 |
| 23 |
259328_at |
AT3G16440
|
ATMLP-300B, myrosinase-binding protein-like protein-300B, MEE36, maternal effect embryo arrest 36, MLP-300B, myrosinase-binding protein-like protein-300B |
0.600 |
-0.185 |
| 24 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.572 |
-0.335 |
| 25 |
253830_at |
AT4G27652
|
unknown |
0.566 |
-0.025 |
| 26 |
253179_at |
AT4G35200
|
unknown |
0.564 |
0.649 |
| 27 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.563 |
0.916 |
| 28 |
245697_at |
AT5G04200
|
AtMC9, metacaspase 9, AtMCP2f, metacaspase 2f, MC9, metacaspase 9, MCP2f, metacaspase 2f |
0.560 |
-0.207 |
| 29 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.536 |
0.579 |
| 30 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.531 |
0.584 |
| 31 |
251178_at |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
0.527 |
0.644 |
| 32 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.517 |
0.715 |
| 33 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
0.498 |
0.506 |
| 34 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
0.495 |
0.548 |
| 35 |
262045_at |
AT1G80240
|
DGR1, DUF642 L-GalL responsive gene 1 |
0.490 |
0.298 |
| 36 |
252765_at |
AT3G42800
|
unknown |
0.481 |
0.792 |
| 37 |
248278_at |
AT5G52890
|
[AT hook motif-containing protein] |
0.471 |
0.604 |
| 38 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
0.469 |
-0.190 |
| 39 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.467 |
0.014 |
| 40 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
0.466 |
0.291 |
| 41 |
248316_at |
AT5G52670
|
[Copper transport protein family] |
0.465 |
0.655 |
| 42 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.464 |
-0.111 |
| 43 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
0.462 |
0.121 |
| 44 |
267409_at |
AT2G34910
|
unknown |
0.460 |
0.140 |
| 45 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
0.453 |
0.615 |
| 46 |
260867_at |
AT1G43790
|
TED6, tracheary element differentiation-related 6 |
0.452 |
-0.105 |
| 47 |
248801_at |
AT5G47370
|
HAT2 |
0.448 |
0.643 |
| 48 |
247869_at |
AT5G57520
|
ATZFP2, ZINC FINGER PROTEIN 2, ZFP2, zinc finger protein 2 |
0.443 |
0.501 |
| 49 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
0.440 |
0.589 |
| 50 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
0.440 |
0.242 |
| 51 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.439 |
0.563 |
| 52 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
0.430 |
1.012 |
| 53 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.429 |
0.099 |
| 54 |
263576_at |
AT2G17080
|
unknown |
0.426 |
0.301 |
| 55 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.424 |
0.403 |
| 56 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.423 |
0.190 |
| 57 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
0.421 |
0.563 |
| 58 |
261777_at |
AT1G76210
|
unknown |
0.420 |
0.343 |
| 59 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.418 |
-0.079 |
| 60 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
0.418 |
0.610 |
| 61 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
0.417 |
0.505 |
| 62 |
254629_at |
AT4G18425
|
unknown |
0.411 |
-0.205 |
| 63 |
253791_at |
AT4G28640
|
IAA11, indole-3-acetic acid inducible 11 |
0.407 |
0.398 |
| 64 |
252557_at |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
0.407 |
0.443 |
| 65 |
245075_at |
AT2G23180
|
CYP96A1, cytochrome P450, family 96, subfamily A, polypeptide 1 |
0.406 |
0.596 |
| 66 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
0.405 |
0.115 |
| 67 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
0.404 |
-0.023 |
| 68 |
264573_at |
AT1G05310
|
[Pectin lyase-like superfamily protein] |
0.403 |
-0.199 |
| 69 |
263931_at |
AT2G36220
|
unknown |
0.401 |
0.222 |
| 70 |
255695_at |
AT4G00080
|
UNE11, unfertilized embryo sac 11 |
0.400 |
0.104 |
| 71 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
0.400 |
0.703 |
| 72 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.396 |
0.166 |
| 73 |
259027_at |
AT3G09280
|
unknown |
0.395 |
0.626 |
| 74 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.390 |
0.056 |
| 75 |
245108_at |
AT2G41510
|
ATCKX1, CYTOKININ OXIDASE/DEHYDROGENASE 1, CKX1, cytokinin oxidase/dehydrogenase 1 |
0.390 |
0.278 |
| 76 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
0.387 |
0.529 |
| 77 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.384 |
0.211 |
| 78 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
0.381 |
0.013 |
| 79 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.380 |
0.070 |
| 80 |
258912_at |
AT3G06460
|
[GNS1/SUR4 membrane protein family] |
0.379 |
-0.194 |
| 81 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
0.377 |
0.130 |
| 82 |
267287_at |
AT2G23630
|
sks16, SKU5 similar 16 |
0.371 |
-0.211 |
| 83 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.371 |
-0.233 |
| 84 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
0.370 |
0.748 |
| 85 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.368 |
-0.071 |
| 86 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.368 |
0.025 |
| 87 |
255028_at |
AT4G09890
|
unknown |
0.366 |
0.411 |
| 88 |
245352_at |
AT4G15490
|
UGT84A3 |
0.365 |
-0.054 |
| 89 |
258183_at |
AT3G21550
|
AtDMP2, Arabidopsis thaliana DUF679 domain membrane protein 2, DMP2, DUF679 domain membrane protein 2 |
0.362 |
-0.161 |
| 90 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
0.362 |
0.401 |
| 91 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
0.361 |
0.213 |
| 92 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.359 |
0.226 |
| 93 |
247177_at |
AT5G65300
|
unknown |
0.359 |
0.159 |
| 94 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.355 |
-0.198 |
| 95 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.354 |
0.640 |
| 96 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.351 |
0.013 |
| 97 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.351 |
0.330 |
| 98 |
257057_at |
AT3G15310
|
unknown |
0.349 |
0.253 |
| 99 |
255074_at |
AT4G09100
|
[RING/U-box superfamily protein] |
0.345 |
0.180 |
| 100 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.343 |
-0.014 |
| 101 |
248286_at |
AT5G52870
|
MAKR5, MEMBRANE-ASSOCIATED KINASE REGULATOR 5 |
0.343 |
0.184 |
| 102 |
267119_at |
AT2G32610
|
ATCSLB01, cellulose synthase-like B1, ATCSLB1, CELLULOSE SYNTHASE LIKE B1, CSLB01, CELLULOSE SYNTHASE LIKE B1, CSLB01, cellulose synthase-like B1 |
0.342 |
-0.080 |
| 103 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
0.341 |
0.109 |
| 104 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.338 |
0.128 |
| 105 |
257026_at |
AT3G19200
|
unknown |
0.337 |
0.340 |
| 106 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.336 |
-0.060 |
| 107 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.336 |
0.179 |
| 108 |
265144_at |
AT1G51170
|
AGC2-3, AGC2 kinase 3, UCN, UNICORN |
0.335 |
0.331 |
| 109 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.333 |
0.399 |
| 110 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
0.332 |
0.206 |
| 111 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.332 |
0.268 |
| 112 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.330 |
0.216 |
| 113 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
0.328 |
-0.034 |
| 114 |
253180_at |
AT4G35210
|
unknown |
0.327 |
0.527 |
| 115 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
0.324 |
-0.136 |
| 116 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
0.323 |
0.058 |
| 117 |
258907_at |
AT3G06370
|
ATNHX4, NHX4, sodium hydrogen exchanger 4 |
0.323 |
0.228 |
| 118 |
246798_at |
AT5G26930
|
GATA23, GATA transcription factor 23 |
0.322 |
0.473 |
| 119 |
251423_at |
AT3G60550
|
CYCP3;2, cyclin p3;2 |
0.322 |
0.166 |
| 120 |
264682_at |
AT1G65570
|
[Pectin lyase-like superfamily protein] |
0.321 |
-0.145 |
| 121 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.321 |
0.122 |
| 122 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
0.320 |
-0.210 |
| 123 |
253044_at |
AT4G37290
|
unknown |
0.319 |
0.164 |
| 124 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.319 |
0.505 |
| 125 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.317 |
-0.088 |
| 126 |
257975_at |
AT3G20830
|
AGC2-4, AGC2 kinase 4, UCNL, UNICORN-like |
0.317 |
0.379 |
| 127 |
253832_at |
AT4G27654
|
unknown |
0.317 |
0.126 |
| 128 |
263575_at |
AT2G17070
|
unknown |
0.316 |
0.241 |
| 129 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
0.315 |
0.360 |
| 130 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
0.314 |
-0.247 |
| 131 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
0.314 |
0.465 |
| 132 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.313 |
0.137 |
| 133 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.312 |
-0.325 |
| 134 |
246534_at |
AT5G15890
|
TBL21, TRICHOME BIREFRINGENCE-LIKE 21 |
0.311 |
-0.008 |
| 135 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
0.308 |
0.789 |
| 136 |
247474_at |
AT5G62280
|
unknown |
0.308 |
0.532 |
| 137 |
254574_at |
AT4G19430
|
unknown |
0.307 |
0.072 |
| 138 |
250327_at |
AT5G12050
|
unknown |
0.306 |
0.495 |
| 139 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
0.305 |
0.035 |
| 140 |
260058_at |
AT1G78100
|
AUF1, auxin up-regulated f-box protein 1 |
0.305 |
0.474 |
| 141 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
0.304 |
0.037 |
| 142 |
255756_at |
AT1G19940
|
AtGH9B5, glycosyl hydrolase 9B5, GH9B5, glycosyl hydrolase 9B5 |
0.303 |
0.071 |
| 143 |
250098_at |
AT5G17350
|
unknown |
0.302 |
-0.001 |
| 144 |
255934_at |
AT1G12740
|
CYP87A2, cytochrome P450, family 87, subfamily A, polypeptide 2 |
0.302 |
0.151 |
| 145 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.302 |
0.036 |
| 146 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
0.302 |
0.142 |
| 147 |
250778_at |
AT5G05500
|
MOP10 |
0.297 |
0.155 |
| 148 |
256190_at |
AT1G30100
|
ATNCED5, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 5, NCED5, nine-cis-epoxycarotenoid dioxygenase 5 |
0.297 |
0.422 |
| 149 |
263282_at |
AT2G14095
|
unknown |
0.296 |
-0.093 |
| 150 |
249522_at |
AT5G38700
|
unknown |
0.295 |
0.176 |
| 151 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
0.294 |
0.065 |
| 152 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
0.292 |
0.049 |
| 153 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
0.291 |
0.121 |
| 154 |
261476_at |
AT1G14480
|
[Ankyrin repeat family protein] |
0.291 |
0.111 |
| 155 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.290 |
0.253 |
| 156 |
245267_at |
AT4G14060
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.289 |
0.030 |
| 157 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.288 |
0.106 |
| 158 |
252204_at |
AT3G50340
|
unknown |
0.288 |
0.342 |
| 159 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.288 |
0.054 |
| 160 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
0.286 |
0.232 |
| 161 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.286 |
0.119 |
| 162 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.285 |
0.127 |
| 163 |
254575_at |
AT4G19460
|
[UDP-Glycosyltransferase superfamily protein] |
0.284 |
-0.003 |
| 164 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.283 |
0.446 |
| 165 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.283 |
0.229 |
| 166 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.281 |
0.070 |
| 167 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
0.280 |
0.079 |
| 168 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.280 |
0.258 |
| 169 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.280 |
0.500 |
| 170 |
250664_at |
AT5G07080
|
[HXXXD-type acyl-transferase family protein] |
0.279 |
-0.080 |
| 171 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
0.278 |
0.174 |
| 172 |
257386_at |
AT2G42440
|
ASL15, ASYMMETRIC LEAVES 2-like 15, LBD17, LOB domain-containing protein 17 |
0.278 |
0.267 |
| 173 |
254158_at |
AT4G24380
|
[INVOLVED IN: 10-formyltetrahydrofolate biosynthetic process, folic acid and derivative biosynthetic process] |
0.276 |
0.071 |
| 174 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.276 |
0.073 |
| 175 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.274 |
0.266 |
| 176 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.274 |
0.436 |
| 177 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.273 |
0.556 |
| 178 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
0.273 |
0.199 |
| 179 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.272 |
0.292 |
| 180 |
262105_at |
AT1G02810
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.271 |
0.118 |
| 181 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
0.269 |
0.174 |
| 182 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.269 |
0.127 |
| 183 |
261367_at |
AT1G53080
|
[Legume lectin family protein] |
0.269 |
0.034 |
| 184 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.269 |
-0.206 |
| 185 |
245172_at |
AT2G47540
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.268 |
-0.101 |
| 186 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
0.268 |
-0.146 |
| 187 |
256779_at |
AT3G13784
|
AtcwINV5, cell wall invertase 5, CWINV5, cell wall invertase 5 |
0.267 |
-0.008 |
| 188 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.267 |
-0.062 |
| 189 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.267 |
-0.087 |
| 190 |
258388_at |
AT3G15370
|
ATEXP12, ATEXPA12, expansin 12, ATHEXP ALPHA 1.24, EXP12, EXPANSIN 12, EXPA12, expansin 12 |
0.267 |
0.130 |
| 191 |
247159_at |
AT5G65800
|
ACS5, ACC synthase 5, ATACS5, CIN5, CYTOKININ-INSENSITIVE 5, ETO2, ETHYLENE OVERPRODUCER 2 |
0.267 |
-0.011 |
| 192 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.266 |
0.081 |
| 193 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.266 |
0.134 |
| 194 |
266346_at |
AT2G01430
|
ATHB-17, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 17, ATHB17, homeobox-leucine zipper protein 17, HB17, homeobox-leucine zipper protein 17 |
0.263 |
0.069 |
| 195 |
246584_at |
AT5G14730
|
unknown |
0.262 |
0.129 |
| 196 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
0.261 |
0.098 |
| 197 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
0.261 |
0.193 |
| 198 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
0.260 |
0.317 |
| 199 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.259 |
0.254 |
| 200 |
258653_at |
AT3G09870
|
SAUR48, SMALL AUXIN UPREGULATED RNA 48 |
0.258 |
0.196 |
| 201 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
0.258 |
0.031 |
| 202 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.258 |
0.111 |
| 203 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
0.257 |
0.476 |
| 204 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.257 |
0.141 |
| 205 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.257 |
0.085 |
| 206 |
256114_at |
AT1G16850
|
unknown |
0.257 |
0.043 |
| 207 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
0.257 |
0.399 |
| 208 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
0.256 |
0.024 |
| 209 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
0.256 |
0.087 |
| 210 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
0.255 |
-0.001 |
| 211 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
0.255 |
0.064 |
| 212 |
252160_at |
AT3G50570
|
[hydroxyproline-rich glycoprotein family protein] |
0.255 |
0.013 |
| 213 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.254 |
-0.030 |
| 214 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.253 |
0.078 |
| 215 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.252 |
0.062 |
| 216 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
0.252 |
0.332 |
| 217 |
248726_at |
AT5G47960
|
ATRABA4C, RAB GTPase homolog A4C, RABA4C, RAB GTPase homolog A4C, SMG1, SMALL MOLECULAR WEIGHT G-PROTEIN 1 |
0.252 |
0.117 |
| 218 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
0.251 |
0.112 |
| 219 |
266756_at |
AT2G46950
|
CYP709B2, cytochrome P450, family 709, subfamily B, polypeptide 2 |
0.251 |
0.103 |
| 220 |
265732_at |
AT2G01300
|
unknown |
0.251 |
0.120 |
| 221 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.250 |
0.057 |
| 222 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.250 |
-0.063 |
| 223 |
267414_at |
AT2G34790
|
EDA28, EMBRYO SAC DEVELOPMENT ARREST 28, MEE23, MATERNAL EFFECT EMBRYO ARREST 23 |
0.250 |
-0.005 |
| 224 |
267623_at |
AT2G39650
|
unknown |
0.250 |
0.059 |
| 225 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
0.247 |
0.238 |
| 226 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
0.246 |
0.530 |
| 227 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
0.243 |
-0.299 |
| 228 |
266831_at |
AT2G22830
|
SQE2, squalene epoxidase 2 |
0.243 |
0.159 |
| 229 |
260227_at |
AT1G74450
|
unknown |
0.243 |
0.017 |
| 230 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
0.243 |
0.010 |
| 231 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
0.242 |
0.092 |
| 232 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.242 |
-0.004 |
| 233 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.242 |
0.031 |
| 234 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
0.242 |
0.118 |
| 235 |
267343_at |
AT2G44260
|
unknown |
0.242 |
-0.105 |
| 236 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.242 |
-0.089 |
| 237 |
247778_at |
AT5G58750
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.241 |
-0.041 |
| 238 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
0.239 |
0.050 |
| 239 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
0.238 |
-0.032 |
| 240 |
249439_at |
AT5G40020
|
[Pathogenesis-related thaumatin superfamily protein] |
0.236 |
-0.178 |
| 241 |
251293_at |
AT3G61930
|
unknown |
0.236 |
0.030 |
| 242 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
0.234 |
0.027 |
| 243 |
267085_at |
AT2G32620
|
ATCSLB02, cellulose synthase-like B, ATCSLB2, CELLULOSE SYNTHASE LIKE B2, CSLB02, CELLULOSE SYNTHASE LIKE B2, CSLB02, cellulose synthase-like B |
0.234 |
-0.179 |
| 244 |
259850_at |
AT1G72240
|
unknown |
0.233 |
0.079 |
| 245 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.233 |
0.034 |
| 246 |
258873_at |
AT3G03240
|
[alpha/beta-Hydrolases superfamily protein] |
0.233 |
0.026 |
| 247 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.233 |
-0.004 |
| 248 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.232 |
0.137 |
| 249 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
0.232 |
0.112 |
| 250 |
265276_at |
AT2G28400
|
unknown |
0.232 |
0.061 |
| 251 |
253405_at |
AT4G32800
|
[Integrase-type DNA-binding superfamily protein] |
0.231 |
0.302 |
| 252 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.231 |
0.091 |
| 253 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.231 |
-0.008 |
| 254 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.230 |
0.209 |
| 255 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-0.891 |
0.011 |
| 256 |
265387_at |
AT2G20670
|
unknown |
-0.764 |
-0.281 |
| 257 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
-0.748 |
0.003 |
| 258 |
261221_at |
AT1G19960
|
unknown |
-0.743 |
0.103 |
| 259 |
261567_at |
AT1G33055
|
unknown |
-0.685 |
-0.310 |
| 260 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
-0.680 |
0.076 |
| 261 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.676 |
-0.715 |
| 262 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.668 |
-0.051 |
| 263 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.655 |
-0.059 |
| 264 |
259385_at |
AT1G13470
|
unknown |
-0.556 |
-0.087 |
| 265 |
258746_at |
AT3G05950
|
[RmlC-like cupins superfamily protein] |
-0.553 |
0.093 |
| 266 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-0.550 |
-0.028 |
| 267 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
-0.519 |
-0.034 |
| 268 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.514 |
0.110 |
| 269 |
254954_at |
AT4G10910
|
unknown |
-0.503 |
-0.080 |
| 270 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
-0.498 |
0.063 |
| 271 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.496 |
-0.006 |
| 272 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.496 |
-0.004 |
| 273 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.487 |
-0.121 |
| 274 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.479 |
0.098 |
| 275 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.479 |
-0.119 |
| 276 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
-0.478 |
-0.030 |
| 277 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
-0.471 |
0.398 |
| 278 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.471 |
0.091 |
| 279 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.465 |
-0.128 |
| 280 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.458 |
0.118 |
| 281 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
-0.457 |
0.041 |
| 282 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.453 |
-0.017 |
| 283 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
-0.453 |
-0.141 |
| 284 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.452 |
-0.158 |
| 285 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
-0.450 |
-0.308 |
| 286 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
-0.443 |
-0.312 |
| 287 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
-0.431 |
-0.181 |
| 288 |
255912_at |
AT1G66960
|
LUP5 |
-0.417 |
-0.011 |
| 289 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.400 |
-0.469 |
| 290 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
-0.396 |
0.469 |
| 291 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
-0.396 |
-0.150 |
| 292 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.394 |
-0.042 |
| 293 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
-0.392 |
0.391 |
| 294 |
254200_at |
AT4G24110
|
unknown |
-0.390 |
-0.562 |
| 295 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.387 |
0.027 |
| 296 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.386 |
-0.010 |
| 297 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
-0.383 |
0.346 |
| 298 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
-0.381 |
0.017 |
| 299 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
-0.379 |
-0.152 |
| 300 |
247024_at |
AT5G66985
|
unknown |
-0.377 |
-0.178 |
| 301 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.376 |
-0.152 |
| 302 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.371 |
0.200 |
| 303 |
259982_at |
AT1G76410
|
ATL8 |
-0.361 |
-0.293 |
| 304 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
-0.356 |
-0.274 |
| 305 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.354 |
0.051 |
| 306 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.353 |
-0.073 |
| 307 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
-0.349 |
0.155 |
| 308 |
255806_at |
AT4G10260
|
[pfkB-like carbohydrate kinase family protein] |
-0.343 |
-0.029 |
| 309 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
-0.340 |
-0.438 |
| 310 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
-0.338 |
0.132 |
| 311 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
-0.336 |
0.092 |
| 312 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
-0.329 |
-0.355 |
| 313 |
247070_at |
AT5G66815
|
unknown |
-0.329 |
-0.354 |
| 314 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.329 |
-0.124 |
| 315 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
-0.328 |
0.024 |
| 316 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.328 |
0.187 |
| 317 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
-0.327 |
0.117 |
| 318 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.325 |
-0.168 |
| 319 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.323 |
-0.038 |
| 320 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
-0.323 |
0.612 |
| 321 |
261198_at |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
-0.320 |
0.069 |
| 322 |
256631_at |
AT3G28320
|
unknown |
-0.317 |
-0.002 |
| 323 |
257473_at |
AT1G33840
|
unknown |
-0.315 |
-0.162 |
| 324 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.315 |
-0.071 |
| 325 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
-0.313 |
-0.066 |
| 326 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.313 |
-0.154 |
| 327 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.310 |
0.087 |
| 328 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
-0.308 |
-0.057 |
| 329 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
-0.307 |
0.066 |
| 330 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
-0.306 |
-0.167 |
| 331 |
254416_at |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
-0.306 |
-0.119 |
| 332 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.298 |
0.041 |
| 333 |
245226_at |
AT3G29970
|
[B12D protein] |
-0.296 |
-0.055 |
| 334 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.296 |
-0.537 |
| 335 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
-0.294 |
-0.311 |
| 336 |
264379_at |
AT2G25200
|
unknown |
-0.290 |
0.146 |
| 337 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
-0.290 |
-0.035 |
| 338 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.289 |
-0.108 |
| 339 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.287 |
0.425 |
| 340 |
256455_at |
AT1G75190
|
unknown |
-0.286 |
0.074 |
| 341 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
-0.286 |
0.087 |
| 342 |
255342_at |
AT4G04510
|
CRK38, cysteine-rich RLK (RECEPTOR-like protein kinase) 38 |
-0.285 |
-0.006 |
| 343 |
263970_at |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
-0.285 |
-0.219 |
| 344 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-0.279 |
0.061 |
| 345 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
-0.278 |
0.152 |
| 346 |
259854_at |
AT1G72200
|
[RING/U-box superfamily protein] |
-0.276 |
-0.066 |
| 347 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.272 |
0.256 |
| 348 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
-0.271 |
-0.095 |
| 349 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.270 |
-0.065 |
| 350 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
-0.269 |
0.234 |
| 351 |
259197_at |
AT3G03670
|
[Peroxidase superfamily protein] |
-0.268 |
0.000 |
| 352 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
-0.264 |
-0.259 |
| 353 |
249941_at |
AT5G22270
|
unknown |
-0.264 |
0.184 |
| 354 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.259 |
0.027 |
| 355 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
-0.257 |
0.081 |
| 356 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
-0.255 |
0.131 |
| 357 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
-0.254 |
0.056 |
| 358 |
252345_at |
AT3G48640
|
unknown |
-0.254 |
0.050 |
| 359 |
248990_at |
AT5G45210
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.252 |
-0.148 |
| 360 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.252 |
-0.154 |
| 361 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
-0.251 |
-0.092 |
| 362 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.250 |
-0.266 |
| 363 |
254894_at |
AT4G11840
|
PLDGAMMA3, phospholipase D gamma 3 |
-0.247 |
0.064 |
| 364 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
-0.247 |
0.010 |
| 365 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.246 |
-0.129 |
| 366 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.246 |
-0.019 |
| 367 |
247991_at |
AT5G56320
|
ATEXP14, ATEXPA14, expansin A14, ATHEXP ALPHA 1.5, EXP14, EXPANSIN 14, EXPA14, expansin A14 |
-0.245 |
-0.465 |
| 368 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.245 |
0.045 |
| 369 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-0.245 |
0.077 |
| 370 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.243 |
-0.130 |
| 371 |
249768_at |
AT5G24100
|
[Leucine-rich repeat protein kinase family protein] |
-0.241 |
-0.313 |
| 372 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
-0.241 |
0.131 |
| 373 |
247585_at |
AT5G60680
|
unknown |
-0.240 |
-0.148 |
| 374 |
261985_at |
AT1G33750
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.240 |
-0.389 |
| 375 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
-0.239 |
0.098 |
| 376 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
-0.238 |
-0.065 |
| 377 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
-0.236 |
-0.125 |
| 378 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
-0.235 |
0.028 |
| 379 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
-0.235 |
-0.027 |
| 380 |
254253_at |
AT4G23320
|
CRK24, cysteine-rich RLK (RECEPTOR-like protein kinase) 24 |
-0.234 |
0.042 |
| 381 |
262170_at |
AT1G74940
|
unknown |
-0.234 |
-0.159 |
| 382 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.233 |
-0.014 |
| 383 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
-0.232 |
-0.114 |
| 384 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.230 |
-0.004 |
| 385 |
259454_at |
AT1G44050
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.229 |
-0.102 |
| 386 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
-0.227 |
0.129 |
| 387 |
264774_at |
AT1G22890
|
unknown |
-0.227 |
0.377 |
| 388 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.227 |
-0.100 |
| 389 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
-0.226 |
0.045 |
| 390 |
253814_at |
AT4G28290
|
unknown |
-0.225 |
-0.093 |
| 391 |
263208_at |
AT1G10480
|
ZFP5, zinc finger protein 5 |
-0.225 |
-0.101 |
| 392 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
-0.224 |
-0.121 |
| 393 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.223 |
-0.008 |
| 394 |
252047_at |
AT3G52490
|
SMXL3, SMAX1-like 3 |
-0.223 |
-0.198 |
| 395 |
253400_at |
AT4G32860
|
unknown |
-0.222 |
-0.272 |
| 396 |
252057_at |
AT3G52480
|
unknown |
-0.222 |
-0.100 |
| 397 |
247948_at |
AT5G57130
|
SMXL5, SMAX1-like 5 |
-0.221 |
0.016 |
| 398 |
256262_at |
AT3G12150
|
unknown |
-0.221 |
-0.047 |
| 399 |
265924_at |
AT2G18620
|
[Terpenoid synthases superfamily protein] |
-0.218 |
-0.158 |
| 400 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
-0.217 |
0.005 |
| 401 |
264836_at |
AT1G03610
|
unknown |
-0.217 |
-0.043 |
| 402 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
-0.216 |
0.062 |
| 403 |
250697_at |
AT5G06800
|
[myb-like HTH transcriptional regulator family protein] |
-0.216 |
-0.135 |
| 404 |
255319_at |
AT4G04220
|
AtRLP46, receptor like protein 46, RLP46, receptor like protein 46 |
-0.216 |
0.093 |
| 405 |
247618_at |
AT5G60280
|
LecRK-I.8, L-type lectin receptor kinase I.8 |
-0.215 |
0.186 |
| 406 |
267226_at |
AT2G44010
|
unknown |
-0.214 |
0.055 |
| 407 |
257925_at |
AT3G23170
|
unknown |
-0.213 |
-0.588 |
| 408 |
259065_at |
AT3G07520
|
ATGLR1.4, GLR1.4, glutamate receptor 1.4 |
-0.211 |
-0.079 |
| 409 |
256980_at |
AT3G26932
|
DRB3, dsRNA-binding protein 3 |
-0.211 |
0.004 |
| 410 |
254571_at |
AT4G19370
|
unknown |
-0.210 |
-0.001 |
| 411 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.209 |
0.250 |
| 412 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.209 |
-0.060 |
| 413 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.208 |
-0.032 |
| 414 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.208 |
-0.089 |
| 415 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
-0.207 |
0.523 |
| 416 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.207 |
0.055 |
| 417 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
-0.207 |
0.077 |
| 418 |
245186_at |
AT1G67710
|
ARR11, response regulator 11 |
-0.207 |
-0.035 |
| 419 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
-0.206 |
0.049 |
| 420 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
-0.205 |
-0.092 |
| 421 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.204 |
-0.108 |
| 422 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.203 |
0.060 |
| 423 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
-0.203 |
-0.250 |
| 424 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
-0.203 |
-0.204 |
| 425 |
262399_at |
AT1G49500
|
unknown |
-0.203 |
-0.017 |
| 426 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
-0.203 |
0.026 |
| 427 |
256358_at |
AT1G66470
|
AtRHD6, RHD6, ROOT HAIR DEFECTIVE6 |
-0.202 |
-0.345 |
| 428 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.202 |
0.154 |
| 429 |
254644_at |
AT4G18510
|
CLE2, CLAVATA3/ESR-related 2 |
-0.201 |
0.005 |
| 430 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
-0.201 |
0.108 |
| 431 |
251857_at |
AT3G54770
|
ARP1, ABA-regulated RNA-binding protein 1 |
-0.198 |
-0.326 |
| 432 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-0.198 |
0.194 |
| 433 |
247754_at |
AT5G59080
|
unknown |
-0.198 |
-0.171 |
| 434 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.197 |
0.097 |
| 435 |
264635_at |
AT1G65500
|
unknown |
-0.197 |
-0.006 |
| 436 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
-0.196 |
0.011 |
| 437 |
253515_at |
AT4G31320
|
SAUR37, SMALL AUXIN UPREGULATED RNA 37 |
-0.196 |
-0.302 |
| 438 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.196 |
-0.057 |
| 439 |
258225_at |
AT3G15630
|
unknown |
-0.196 |
0.046 |
| 440 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
-0.196 |
-0.174 |
| 441 |
249384_at |
AT5G39890
|
unknown |
-0.195 |
0.012 |
| 442 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.194 |
0.066 |
| 443 |
255858_at |
AT1G67030
|
ZFP6, zinc finger protein 6 |
-0.191 |
-0.141 |
| 444 |
251419_at |
AT3G60470
|
unknown |
-0.190 |
0.073 |
| 445 |
252882_at |
AT4G39675
|
unknown |
-0.190 |
-0.102 |
| 446 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.190 |
0.187 |
| 447 |
259737_at |
AT1G64400
|
LACS3, long-chain acyl-CoA synthetase 3 |
-0.189 |
0.195 |
| 448 |
252352_at |
AT3G48185
|
unknown |
-0.189 |
-0.041 |
| 449 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.188 |
-0.136 |
| 450 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
-0.188 |
-0.108 |
| 451 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.186 |
-0.153 |
| 452 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
-0.186 |
0.030 |
| 453 |
249824_at |
AT5G23380
|
unknown |
-0.186 |
0.070 |
| 454 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
-0.186 |
-0.045 |
| 455 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-0.186 |
0.133 |
| 456 |
255957_at |
AT1G22160
|
unknown |
-0.184 |
0.165 |
| 457 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.184 |
0.124 |
| 458 |
250640_at |
AT5G07150
|
[Leucine-rich repeat protein kinase family protein] |
-0.182 |
0.109 |
| 459 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.181 |
0.020 |
| 460 |
249972_at |
AT5G19040
|
ATIPT5, Arabidopsis thaliana ISOPENTENYLTRANSFERASE 5, IPT5, isopentenyltransferase 5 |
-0.181 |
-0.154 |
| 461 |
249709_at |
AT5G35670
|
iqd33, IQ-domain 33 |
-0.180 |
-0.182 |
| 462 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
-0.180 |
0.015 |
| 463 |
246216_at |
AT4G36380
|
ROT3, ROTUNDIFOLIA 3 |
-0.180 |
-0.047 |
| 464 |
265611_at |
AT2G25510
|
unknown |
-0.179 |
-0.039 |
| 465 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-0.178 |
-0.024 |
| 466 |
263490_at |
AT2G42620
|
MAX2, MORE AXILLARY BRANCHES 2, ORE9, ORESARA 9, PPS, PLEIOTROPIC PHOTOSIGNALING |
-0.178 |
-0.026 |
| 467 |
254175_at |
AT4G24050
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.178 |
-0.167 |
| 468 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.177 |
0.137 |
| 469 |
250152_at |
AT5G15120
|
unknown |
-0.177 |
-0.022 |
| 470 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
-0.177 |
-0.045 |
| 471 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
-0.176 |
-0.187 |
| 472 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.176 |
-0.096 |
| 473 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
-0.176 |
0.298 |
| 474 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
-0.175 |
-0.134 |
| 475 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.175 |
-0.121 |
| 476 |
247829_at |
AT5G58520
|
[Protein kinase superfamily protein] |
-0.175 |
0.026 |
| 477 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.175 |
0.035 |
| 478 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.174 |
-0.355 |
| 479 |
254797_at |
AT4G13030
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.173 |
0.088 |
| 480 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
-0.172 |
0.012 |
| 481 |
263265_at |
AT2G38820
|
unknown |
-0.172 |
0.002 |
| 482 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
-0.172 |
-0.180 |
| 483 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
-0.172 |
0.031 |
| 484 |
248981_at |
AT5G45110
|
ATNPR3, NPR3, NPR1-like protein 3 |
-0.172 |
-0.000 |
| 485 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
-0.171 |
-0.358 |
| 486 |
259976_at |
AT1G76560
|
CP12-3, CP12 domain-containing protein 3 |
-0.171 |
0.102 |
| 487 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
-0.171 |
-0.065 |
| 488 |
266658_at |
AT2G25735
|
unknown |
-0.171 |
-0.056 |
| 489 |
263032_at |
AT1G23850
|
unknown |
-0.170 |
0.021 |
| 490 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
-0.170 |
0.071 |
| 491 |
248092_at |
AT5G55170
|
ATSUMO3, SUM3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO 3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO3, small ubiquitin-like modifier 3 |
-0.170 |
0.082 |
| 492 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.170 |
-0.058 |
| 493 |
259853_at |
AT1G72300
|
PSY1R, PSY1 receptor |
-0.169 |
0.069 |
| 494 |
260211_at |
AT1G74440
|
unknown |
-0.168 |
0.066 |
| 495 |
260303_at |
AT1G70520
|
ASG6, ALTERED SEED GERMINATION 6, CRK2, cysteine-rich RLK (RECEPTOR-like protein kinase) 2 |
-0.167 |
0.092 |
| 496 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.167 |
-0.260 |
| 497 |
260754_at |
AT1G49000
|
unknown |
-0.167 |
0.107 |
| 498 |
250747_at |
AT5G05900
|
[UDP-Glycosyltransferase superfamily protein] |
-0.166 |
-0.043 |
| 499 |
266259_at |
AT2G27830
|
unknown |
-0.165 |
-0.134 |
| 500 |
254852_at |
AT4G12020
|
ATWRKY19, MAPKKK11, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 11, MEKK4, MAPK/ERK KINASE KINASE 4, WRKY19 |
-0.165 |
0.144 |
| 501 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.165 |
-0.204 |
| 502 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
-0.164 |
0.233 |
| 503 |
253886_at |
AT4G27710
|
CYP709B3, cytochrome P450, family 709, subfamily B, polypeptide 3 |
-0.164 |
0.140 |
| 504 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
-0.164 |
-0.112 |
| 505 |
245602_at |
AT4G14270
|
unknown |
-0.164 |
-0.029 |
| 506 |
246911_at |
AT5G25810
|
tny, TINY |
-0.164 |
-0.144 |
| 507 |
246366_at |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
-0.163 |
-0.017 |
| 508 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
-0.163 |
-0.137 |