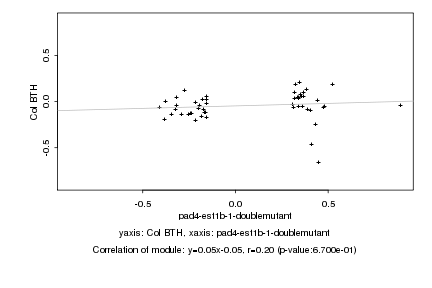
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pad4-est1b-1-doublemutant |
Log2 signal ratio
Col BTH |
| 1 |
246394_at |
AT1G58160
|
JAX1, JACALIN-TYPE LECTIN REQUIRED FOR POTEXVIRUS RESISTANCE 1 |
0.888 |
-0.034 |
| 2 |
246951_at |
AT5G04880
|
[expressed protein] |
0.519 |
0.190 |
| 3 |
257475_at |
AT1G80880
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.475 |
-0.044 |
| 4 |
262619_at |
AT1G06550
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.473 |
-0.055 |
| 5 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.443 |
-0.650 |
| 6 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.442 |
0.012 |
| 7 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
0.429 |
-0.239 |
| 8 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
0.406 |
-0.456 |
| 9 |
250214_at |
AT5G13870
|
EXGT-A4, endoxyloglucan transferase A4, XTH5, xyloglucan endotransglucosylase/hydrolase 5 |
0.403 |
-0.087 |
| 10 |
256741_at |
AT3G29375
|
[XH domain-containing protein] |
0.384 |
-0.082 |
| 11 |
267397_at |
AT1G76170
|
[2-thiocytidine tRNA biosynthesis protein, TtcA] |
0.378 |
0.138 |
| 12 |
251100_at |
AT5G01670
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.364 |
0.057 |
| 13 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
0.363 |
0.106 |
| 14 |
252241_at |
AT3G50050
|
[Eukaryotic aspartyl protease family protein] |
0.357 |
-0.050 |
| 15 |
261559_at |
AT1G01780
|
PLIM2b, PLIM2b |
0.351 |
0.057 |
| 16 |
257655_at |
AT3G13350
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
0.346 |
0.083 |
| 17 |
263854_at |
AT2G04430
|
atnudt5, nudix hydrolase homolog 5, NUDT5, nudix hydrolase homolog 5 |
0.345 |
0.211 |
| 18 |
249824_at |
AT5G23380
|
unknown |
0.339 |
0.050 |
| 19 |
265302_at |
AT2G13970
|
[Mutator-like transposase family, has a 4.1e-39 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.338 |
0.043 |
| 20 |
249640_at |
AT5G36940
|
CAT3, cationic amino acid transporter 3 |
0.335 |
-0.046 |
| 21 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
0.330 |
0.047 |
| 22 |
256930_at |
AT3G22460
|
OASA2, O-acetylserine (thiol) lyase (OAS-TL) isoform A2 |
0.321 |
0.195 |
| 23 |
263437_at |
AT2G28670
|
ESB1, ENHANCED SUBERIN 1 |
0.318 |
0.035 |
| 24 |
254948_at |
AT4G11000
|
[Ankyrin repeat family protein] |
0.315 |
0.105 |
| 25 |
245770_at |
AT1G30240
|
[FUNCTIONS IN: binding] |
0.309 |
-0.064 |
| 26 |
257616_at |
AT3G26540
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.305 |
-0.027 |
| 27 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.413 |
-0.062 |
| 28 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.383 |
-0.194 |
| 29 |
246066_at |
AT5G19400
|
SMG7 |
-0.381 |
0.011 |
| 30 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
-0.350 |
-0.131 |
| 31 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.325 |
-0.080 |
| 32 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.323 |
0.053 |
| 33 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.320 |
-0.033 |
| 34 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.292 |
-0.130 |
| 35 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.277 |
0.126 |
| 36 |
251750_at |
AT3G55710
|
[UDP-Glycosyltransferase superfamily protein] |
-0.257 |
-0.139 |
| 37 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
-0.247 |
-0.128 |
| 38 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.242 |
-0.126 |
| 39 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
-0.220 |
-0.201 |
| 40 |
265323_at |
AT2G18260
|
ATSYP112, SYP112, syntaxin of plants 112 |
-0.219 |
-0.002 |
| 41 |
254996_at |
AT4G10390
|
[Protein kinase superfamily protein] |
-0.203 |
-0.069 |
| 42 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
-0.194 |
-0.038 |
| 43 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.188 |
-0.161 |
| 44 |
249051_at |
AT5G44390
|
[FAD-binding Berberine family protein] |
-0.179 |
0.031 |
| 45 |
257051_at |
AT3G15270
|
SPL5, squamosa promoter binding protein-like 5 |
-0.175 |
-0.085 |
| 46 |
246142_at |
AT5G19970
|
unknown |
-0.171 |
-0.117 |
| 47 |
247118_at |
AT5G65890
|
ACR1, ACT domain repeat 1 |
-0.163 |
-0.114 |
| 48 |
257322_at |
ATMG01180
|
ORF111B |
-0.161 |
0.031 |
| 49 |
254235_at |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
-0.160 |
-0.021 |
| 50 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.159 |
0.059 |
| 51 |
259373_at |
AT1G69160
|
unknown |
-0.159 |
-0.168 |