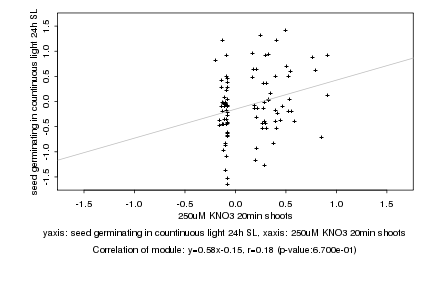
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
250uM KNO3 20min shoots |
Log2 signal ratio
seed germinating in countinuous light 24h SL |
| 1 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
0.908 |
0.933 |
| 2 |
262180_at |
AT1G78050
|
PGM, phosphoglycerate/bisphosphoglycerate mutase |
0.904 |
0.127 |
| 3 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.848 |
-0.707 |
| 4 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
0.785 |
0.620 |
| 5 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.756 |
0.887 |
| 6 |
259015_at |
AT3G07350
|
unknown |
0.582 |
-0.384 |
| 7 |
262284_at |
AT1G68670
|
[myb-like transcription factor family protein] |
0.554 |
-0.185 |
| 8 |
249325_at |
AT5G40850
|
UPM1, urophorphyrin methylase 1 |
0.537 |
0.608 |
| 9 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
0.531 |
0.054 |
| 10 |
247754_at |
AT5G59080
|
unknown |
0.519 |
-0.198 |
| 11 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
0.517 |
0.509 |
| 12 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.503 |
0.714 |
| 13 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.494 |
1.427 |
| 14 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.462 |
-0.099 |
| 15 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
0.445 |
-0.361 |
| 16 |
245977_at |
AT5G13110
|
G6PD2, glucose-6-phosphate dehydrogenase 2 |
0.413 |
-0.227 |
| 17 |
249266_at |
AT5G41670
|
[6-phosphogluconate dehydrogenase family protein] |
0.402 |
1.231 |
| 18 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
0.399 |
-0.533 |
| 19 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.393 |
-0.394 |
| 20 |
246981_at |
AT5G04840
|
[bZIP protein] |
0.388 |
-0.169 |
| 21 |
258437_at |
AT3G16560
|
[Protein phosphatase 2C family protein] |
0.387 |
0.498 |
| 22 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
0.376 |
-0.819 |
| 23 |
260623_at |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
0.339 |
0.174 |
| 24 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
0.325 |
0.028 |
| 25 |
248253_at |
AT5G53290
|
CRF3, cytokinin response factor 3 |
0.324 |
0.942 |
| 26 |
254235_at |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
0.323 |
0.048 |
| 27 |
256528_at |
AT1G66140
|
ZFP4, zinc finger protein 4 |
0.305 |
-0.533 |
| 28 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
0.300 |
0.373 |
| 29 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
0.294 |
-0.421 |
| 30 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
0.291 |
0.922 |
| 31 |
260771_at |
AT1G49160
|
WNK7 |
0.286 |
-1.255 |
| 32 |
248125_at |
AT5G54740
|
SESA5, seed storage albumin 5 |
0.285 |
-0.006 |
| 33 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.283 |
-0.397 |
| 34 |
264790_at |
AT2G17820
|
AHK1, ATHK1, histidine kinase 1, HK1, histidine kinase 1 |
0.271 |
-0.136 |
| 35 |
248199_at |
AT5G54170
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.270 |
0.373 |
| 36 |
249359_at |
AT5G40470
|
[RNI-like superfamily protein] |
0.263 |
-0.421 |
| 37 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
0.261 |
-0.528 |
| 38 |
260325_at |
AT1G63940
|
MDAR6, monodehydroascorbate reductase 6 |
0.243 |
1.323 |
| 39 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
0.210 |
-0.123 |
| 40 |
262323_at |
AT1G64190
|
[6-phosphogluconate dehydrogenase family protein] |
0.207 |
0.655 |
| 41 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
0.203 |
-0.926 |
| 42 |
250597_at |
AT5G07680
|
ANAC079, Arabidopsis NAC domain containing protein 79, ANAC080, NAC domain containing protein 80, ATNAC4, NAC080, NAC domain containing protein 80, NAC4 |
0.200 |
-0.314 |
| 43 |
250230_at |
AT5G13900
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.192 |
-1.167 |
| 44 |
261195_at |
AT1G12820
|
AFB3, auxin signaling F-box 3 |
0.183 |
-0.127 |
| 45 |
258350_at |
AT3G17510
|
CIPK1, CBL-interacting protein kinase 1, SnRK3.16, SNF1-RELATED PROTEIN KINASE 3.16 |
0.180 |
-0.062 |
| 46 |
252407_at |
AT3G47520
|
MDH, malate dehydrogenase, pNAD-MDH, plastidic NAD-dependent malate dehydrogenase |
0.178 |
0.641 |
| 47 |
261193_at |
AT1G32920
|
unknown |
0.164 |
0.488 |
| 48 |
263731_at |
AT1G59970
|
[Matrixin family protein] |
0.163 |
0.965 |
| 49 |
248205_at |
AT5G54300
|
unknown |
-0.203 |
0.820 |
| 50 |
260561_at |
AT2G43580
|
[Chitinase family protein] |
-0.164 |
-0.468 |
| 51 |
255271_at |
AT4G05260
|
[Ubiquitin-like superfamily protein] |
-0.158 |
-0.367 |
| 52 |
247649_at |
AT5G60030
|
unknown |
-0.143 |
0.420 |
| 53 |
264583_at |
AT1G05170
|
[Galactosyltransferase family protein] |
-0.141 |
-0.086 |
| 54 |
261575_at |
AT1G01130
|
unknown |
-0.139 |
0.280 |
| 55 |
263291_at |
AT2G10920
|
unknown |
-0.137 |
-0.005 |
| 56 |
267032_at |
AT2G38490
|
CIPK22, CBL-interacting protein kinase 22, SnRK3.19, SNF1-RELATED PROTEIN KINASE 3.19 |
-0.135 |
-0.451 |
| 57 |
260395_at |
AT1G69780
|
ATHB13 |
-0.133 |
-0.186 |
| 58 |
246001_at |
AT5G20790
|
unknown |
-0.132 |
1.230 |
| 59 |
265862_at |
AT2G01780
|
[Curculin-like (mannose-binding) lectin family protein] |
-0.129 |
-0.446 |
| 60 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
-0.123 |
-0.449 |
| 61 |
256562_at |
AT3G29775
|
[copia-like retrotransposon family, has a 2.8e-246 P-value blast match to gb|AAO73527.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.122 |
-0.963 |
| 62 |
267160_at |
AT2G37670
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.119 |
-0.020 |
| 63 |
261247_at |
AT1G20070
|
unknown |
-0.117 |
-0.094 |
| 64 |
260327_at |
AT1G63840
|
[RING/U-box superfamily protein] |
-0.113 |
0.095 |
| 65 |
258367_at |
AT3G14370
|
WAG2 |
-0.113 |
-0.059 |
| 66 |
249511_at |
AT5G38500
|
[FUNCTIONS IN: DNA binding] |
-0.110 |
-0.349 |
| 67 |
262384_at |
AT1G72950
|
[Disease resistance protein (TIR-NBS class)] |
-0.104 |
-0.824 |
| 68 |
266619_at |
AT2G35460
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.103 |
-0.856 |
| 69 |
265099_at |
AT1G03990
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.100 |
-0.032 |
| 70 |
261024_at |
AT1G12190
|
[F-box and associated interaction domains-containing protein] |
-0.100 |
-1.360 |
| 71 |
261812_at |
AT1G08270
|
unknown |
-0.099 |
-0.004 |
| 72 |
258021_at |
AT3G19380
|
PUB25, plant U-box 25 |
-0.097 |
-0.357 |
| 73 |
262996_at |
AT1G54440
|
[Polynucleotidyl transferase, ribonuclease H fold protein with HRDC domain] |
-0.095 |
0.920 |
| 74 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
-0.095 |
-0.066 |
| 75 |
264181_at |
AT1G65350
|
UBQ13, ubiquitin 13 |
-0.094 |
-0.457 |
| 76 |
258192_at |
AT3G29110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.094 |
0.236 |
| 77 |
258012_at |
AT3G19310
|
[PLC-like phosphodiesterases superfamily protein] |
-0.093 |
-1.087 |
| 78 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.092 |
0.517 |
| 79 |
265483_at |
AT2G15790
|
CYP40, CYCLOPHILIN 40, SQN, SQUINT |
-0.091 |
-0.166 |
| 80 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.088 |
0.459 |
| 81 |
245419_at |
AT4G17380
|
ATMSH4, ARABIDOPSIS MUTS HOMOLOG 4, MSH4, MUTS HOMOLOG 4, MSH4, MUTS-like protein 4 |
-0.088 |
-0.210 |
| 82 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
-0.088 |
-0.062 |
| 83 |
246125_at |
AT5G19875
|
unknown |
-0.087 |
-0.673 |
| 84 |
267557_at |
AT2G32710
|
ACK2, CYCLIN-DEPENDENT KINASE INHIBITOR 2, ICK7, INTERACTORS OF CDC2 KINASE 7, KRP4, KRP4, KIP-RELATED PROTEIN 4 |
-0.087 |
-0.400 |
| 85 |
259086_at |
AT3G04990
|
unknown |
-0.087 |
-1.631 |
| 86 |
260058_at |
AT1G78100
|
AUF1, auxin up-regulated f-box protein 1 |
-0.087 |
0.393 |
| 87 |
258385_at |
AT3G15510
|
ANAC056, Arabidopsis NAC domain containing protein 56, ATNAC2, NAC domain containing protein 2, NAC2, NAC domain containing protein 2, NARS1, NAC-REGULATED SEED MORPHOLOGY 1 |
-0.086 |
-0.426 |
| 88 |
255772_at |
AT1G18530
|
[EF hand calcium-binding protein family] |
-0.086 |
-0.259 |
| 89 |
249937_at |
AT5G22470
|
[NAD+ ADP-ribosyltransferases] |
-0.086 |
0.054 |
| 90 |
251088_at |
AT5G01480
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.086 |
-0.625 |
| 91 |
248661_at |
AT5G48670
|
AGL80, AGAMOUS-like 80, FEM111 |
-0.086 |
-0.613 |
| 92 |
263423_at |
AT2G31700
|
unknown |
-0.085 |
-1.526 |
| 93 |
245107_at |
AT2G41690
|
AT-HSFB3, heat shock transcription factor B3, HSFB3, HEAT SHOCK TRANSCRIPTION FACTOR B3, HSFB3, heat shock transcription factor B3 |
-0.085 |
-0.690 |
| 94 |
259617_at |
AT1G47970
|
unknown |
-0.083 |
-0.083 |
| 95 |
246332_at |
AT3G44830
|
[Lecithin:cholesterol acyltransferase family protein] |
-0.083 |
0.290 |