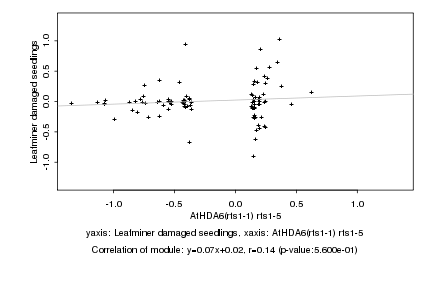
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
AtHDA6(rts1-1) rts1-5 |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.623 |
0.159 |
| 2 |
259252_at |
AT3G07610
|
IBM1, increase in bonsai methylation 1 |
0.458 |
-0.044 |
| 3 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
0.377 |
0.248 |
| 4 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.360 |
1.035 |
| 5 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.342 |
0.656 |
| 6 |
256569_at |
AT3G19550
|
unknown |
0.272 |
0.577 |
| 7 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
0.259 |
0.384 |
| 8 |
260627_at |
AT1G62310
|
[transcription factor jumonji (jmjC) domain-containing protein] |
0.243 |
0.301 |
| 9 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
0.241 |
0.008 |
| 10 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.240 |
-0.412 |
| 11 |
253910_at |
AT4G27290
|
[S-locus lectin protein kinase family protein] |
0.238 |
-0.007 |
| 12 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.237 |
0.415 |
| 13 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
0.235 |
-0.404 |
| 14 |
266968_at |
AT2G39360
|
[Protein kinase superfamily protein] |
0.229 |
0.128 |
| 15 |
252160_at |
AT3G50570
|
[hydroxyproline-rich glycoprotein family protein] |
0.209 |
-0.258 |
| 16 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.201 |
0.866 |
| 17 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
0.196 |
-0.432 |
| 18 |
262552_at |
AT1G31350
|
KUF1, KAR-UP F-box 1 |
0.195 |
0.038 |
| 19 |
260909_at |
AT1G02670
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.193 |
0.016 |
| 20 |
247619_at |
AT5G60290
|
unknown |
0.193 |
-0.033 |
| 21 |
261924_at |
AT1G22550
|
[Major facilitator superfamily protein] |
0.192 |
0.077 |
| 22 |
245477_at |
AT4G16110
|
ARR2, response regulator 2, RR2, response regulator 2 |
0.189 |
-0.042 |
| 23 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
0.187 |
-0.380 |
| 24 |
246397_at |
AT1G58190
|
AtRLP9, receptor like protein 9, RLP9, receptor like protein 9 |
0.182 |
0.034 |
| 25 |
266175_at |
AT2G02450
|
ANAC034, Arabidopsis NAC domain containing protein 34, ANAC035, NAC domain containing protein 35, AtLOV1, LOV1, LONG VEGETATIVE PHASE 1, NAC035, NAC domain containing protein 35 |
0.175 |
0.328 |
| 26 |
248205_at |
AT5G54300
|
unknown |
0.171 |
0.559 |
| 27 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
0.165 |
-0.467 |
| 28 |
267148_at |
AT2G23470
|
RUS4, ROOT UV-B SENSITIVE 4 |
0.162 |
-0.253 |
| 29 |
255037_at |
AT4G09460
|
AtMYB6, myb domain protein 6, MYB6, myb domain protein 6 |
0.160 |
0.067 |
| 30 |
256674_at |
AT3G52360
|
unknown |
0.157 |
-0.613 |
| 31 |
264519_at |
AT1G10000
|
[Ribonuclease H-like superfamily protein] |
0.156 |
-0.037 |
| 32 |
260970_at |
AT1G53640
|
unknown |
0.156 |
0.012 |
| 33 |
266118_at |
AT2G02130
|
LCR68, low-molecular-weight cysteine-rich 68, PDF2.3 |
0.154 |
-0.273 |
| 34 |
259853_at |
AT1G72300
|
PSY1R, PSY1 receptor |
0.151 |
-0.228 |
| 35 |
260641_at |
AT1G53200
|
unknown |
0.149 |
-0.095 |
| 36 |
252298_at |
AT3G49060
|
[U-box domain-containing protein kinase family protein] |
0.149 |
-0.107 |
| 37 |
264663_at |
AT1G09970
|
LRR XI-23, RLK7, receptor-like kinase 7 |
0.149 |
0.344 |
| 38 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.146 |
0.287 |
| 39 |
246524_at |
AT5G15860
|
ATPCME, prenylcysteine methylesterase, ICME, Isoprenylcysteine methylesterase, PCME, prenylcysteine methylesterase |
0.143 |
-0.263 |
| 40 |
267225_at |
AT2G44000
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.141 |
0.045 |
| 41 |
267581_at |
AT2G41980
|
[Protein with RING/U-box and TRAF-like domains] |
0.141 |
-0.100 |
| 42 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.140 |
-0.892 |
| 43 |
250084_at |
AT5G17240
|
SDG40, SET domain group 40 |
0.138 |
-0.112 |
| 44 |
248135_at |
AT5G54890
|
mCSF2, mitochondrial CAF-like splicing factor 2 |
0.135 |
-0.089 |
| 45 |
248936_at |
AT5G45710
|
AT-HSFA4C, HEAT SHOCK TRANSCRIPTION FACTOR A4C, HSFA4C, HEAT SHOCK TRANSCRIPTION FACTOR A4C, RHA1, ROOT HANDEDNESS 1 |
0.134 |
0.110 |
| 46 |
252144_at |
AT3G51190
|
[Ribosomal protein L2 family] |
0.132 |
-0.014 |
| 47 |
267632_at |
AT2G42160
|
BRIZ1, BRAP2 RING ZnF UBP domain-containing protein 1 |
0.130 |
0.119 |
| 48 |
252979_at |
AT4G38225
|
unknown |
0.129 |
-0.075 |
| 49 |
248949_at |
AT5G45570
|
[Ulp1 protease family protein] |
-1.351 |
-0.024 |
| 50 |
263675_x_at |
AT2G04770
|
[CACTA-like transposase family (Ptta/En/Spm), has a 6.0e-20 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-1.133 |
0.000 |
| 51 |
249259_at |
AT5G41660
|
unknown |
-1.080 |
-0.036 |
| 52 |
263479_x_at |
AT2G04000
|
unknown |
-1.075 |
-0.032 |
| 53 |
250605_at |
AT5G07570
|
[glycine/proline-rich protein] |
-1.070 |
0.022 |
| 54 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.995 |
-0.287 |
| 55 |
250979_at |
AT5G03090
|
unknown |
-0.874 |
0.000 |
| 56 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
-0.849 |
-0.132 |
| 57 |
265380_at |
AT2G16670
|
[copia-like retrotransposon family, has a 1.2e-190 P-value blast match to GB:AAB82754 retrofit (TY1_Copia-element) (Oryza longistaminata)] |
-0.821 |
0.010 |
| 58 |
249726_at |
AT5G35480
|
unknown |
-0.809 |
-0.171 |
| 59 |
266393_at |
AT2G41260
|
ATM17, M17 |
-0.783 |
0.037 |
| 60 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
-0.769 |
-0.004 |
| 61 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
-0.759 |
0.090 |
| 62 |
246888_at |
AT5G26270
|
unknown |
-0.747 |
0.273 |
| 63 |
262719_at |
AT1G43590
|
unknown |
-0.742 |
-0.017 |
| 64 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.720 |
-0.247 |
| 65 |
250135_at |
AT5G15360
|
unknown |
-0.646 |
-0.011 |
| 66 |
253713_at |
AT4G29370
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.640 |
-0.013 |
| 67 |
249406_at |
AT5G40210
|
UMAMIT42, Usually multiple acids move in and out Transporters 42 |
-0.631 |
0.354 |
| 68 |
252663_at |
AT3G44070
|
[Glycosyl hydrolase family 35 protein] |
-0.627 |
0.002 |
| 69 |
263851_at |
AT2G04460
|
unknown |
-0.625 |
-0.233 |
| 70 |
245032_at |
AT2G26630
|
[transposase IS4 family protein, contains Pfam profile: PF01609 transposase DDE domain] |
-0.595 |
-0.059 |
| 71 |
246390_at |
AT1G77330
|
ACO5, ACC oxidase 5 |
-0.557 |
-0.124 |
| 72 |
263367_at |
AT2G20460
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-0.555 |
0.044 |
| 73 |
257354_x_at |
AT2G23480
|
unknown |
-0.551 |
0.005 |
| 74 |
257284_at |
AT3G29650
|
[CACTA-like transposase family (Tnp1/En/Spm), has a 5.1e-81 P-value blast match to ref|NP_189784.1| TNP1-related protein (Arabidopsis thaliana) (CACTA-element)] |
-0.543 |
-0.005 |
| 75 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.538 |
-0.003 |
| 76 |
248394_at |
AT5G52070
|
[Agenet domain-containing protein] |
-0.532 |
0.012 |
| 77 |
255376_x_at |
AT4G03790
|
[gypsy-like retrotransposon family (Athila), has a 2.2e-286 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-0.531 |
-0.038 |
| 78 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.466 |
0.322 |
| 79 |
262031_x_at |
AT1G37160
|
[gypsy-like retrotransposon family (Athila), similar to putative Athila retroelement ORF1 protein GI:4567296 from (Arabidopsis thaliana)] |
-0.438 |
-0.002 |
| 80 |
249715_at |
AT5G35760
|
[Beta-galactosidase related protein] |
-0.431 |
-0.020 |
| 81 |
252545_at |
AT3G45820
|
unknown |
-0.430 |
0.018 |
| 82 |
256460_at |
AT1G36240
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
-0.425 |
-0.079 |
| 83 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
-0.420 |
-0.034 |
| 84 |
265503_at |
AT2G15510
|
[non-LTR retrotransposon family (LINE), has a 3.0e-20 P-value blast match to GB:NP_038602 L1 repeat, Tf subfamily, member 18 (LINE-element) (Mus musculus)] |
-0.418 |
0.025 |
| 85 |
256602_at |
AT3G28310
|
unknown |
-0.417 |
0.014 |
| 86 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.411 |
-0.094 |
| 87 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.410 |
0.951 |
| 88 |
250639_at |
AT5G07560
|
ATGRP20, GRP20, glycine-rich protein 20 |
-0.409 |
0.098 |
| 89 |
247640_at |
AT5G60610
|
[F-box/RNI-like superfamily protein] |
-0.401 |
-0.068 |
| 90 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.384 |
-0.673 |
| 91 |
245103_at |
AT2G41590
|
unknown |
-0.383 |
0.049 |
| 92 |
262615_at |
AT1G13950
|
ATELF5A-1, EUKARYOTIC ELONGATION FACTOR 5A-1, EIF-5A, EIF5A, EUKARYOTIC ELONGATION FACTOR 5A, ELF5A-1, eukaryotic elongation factor 5A-1 |
-0.380 |
0.064 |
| 93 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
-0.374 |
-0.063 |
| 94 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.366 |
-0.129 |
| 95 |
248015_at |
AT5G56370
|
[F-box/RNI-like/FBD-like domains-containing protein] |
-0.366 |
-0.016 |