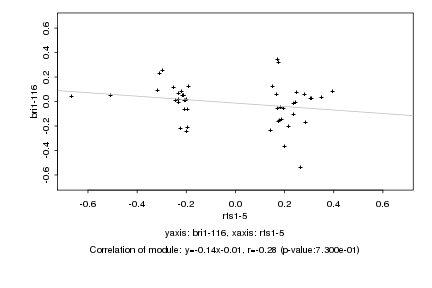
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
rts1-5 |
Log2 signal ratio
bri1-116 |
| 1 |
246698_at |
AT5G30480
|
[CACTA-like transposase family (Ptta/En/Spm), has a 6.2e-37 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.393 |
0.084 |
| 2 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
0.349 |
0.033 |
| 3 |
266151_x_at |
AT2G12300
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.8e-41 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.309 |
0.029 |
| 4 |
255463_at |
AT4G02960
|
ATRE2, retro element 2, RE2, retro element 2 |
0.302 |
0.025 |
| 5 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
0.284 |
-0.166 |
| 6 |
256569_at |
AT3G19550
|
unknown |
0.278 |
0.058 |
| 7 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.264 |
-0.538 |
| 8 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.248 |
0.078 |
| 9 |
257551_at |
AT3G13270
|
unknown |
0.243 |
-0.000 |
| 10 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.233 |
-0.014 |
| 11 |
255460_at |
AT4G02800
|
unknown |
0.233 |
-0.101 |
| 12 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
0.212 |
-0.203 |
| 13 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
0.199 |
-0.360 |
| 14 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
0.192 |
-0.055 |
| 15 |
247116_at |
AT5G65970
|
ATMLO10, MILDEW RESISTANCE LOCUS O 10, MLO10, MILDEW RESISTANCE LOCUS O 10 |
0.184 |
-0.146 |
| 16 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
0.183 |
-0.044 |
| 17 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
0.176 |
-0.150 |
| 18 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
0.175 |
0.322 |
| 19 |
261224_at |
AT1G20160
|
ATSBT5.2 |
0.172 |
-0.162 |
| 20 |
258059_at |
AT3G29035
|
ANAC059, Arabidopsis NAC domain containing protein 59, ATNAC3, NAC domain containing protein 3, NAC3, NAC domain containing protein 3, ORS1, ORE1 SISTER1 |
0.171 |
-0.050 |
| 21 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
0.171 |
0.345 |
| 22 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
0.163 |
0.064 |
| 23 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
0.150 |
0.126 |
| 24 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
0.141 |
-0.232 |
| 25 |
249406_at |
AT5G40210
|
UMAMIT42, Usually multiple acids move in and out Transporters 42 |
-0.670 |
0.042 |
| 26 |
255376_x_at |
AT4G03790
|
[gypsy-like retrotransposon family (Athila), has a 2.2e-286 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-0.509 |
0.052 |
| 27 |
263367_at |
AT2G20460
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-0.318 |
0.095 |
| 28 |
253713_at |
AT4G29370
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.309 |
0.233 |
| 29 |
249425_at |
AT5G39790
|
5'-AMP-activated protein kinase-related |
-0.297 |
0.254 |
| 30 |
259123_at |
AT3G02200
|
[Proteasome component (PCI) domain protein] |
-0.253 |
0.118 |
| 31 |
249471_at |
AT5G39360
|
EDL2, EID1-like 2 |
-0.245 |
0.015 |
| 32 |
249464_at |
AT5G39710
|
EMB2745, EMBRYO DEFECTIVE 2745 |
-0.235 |
-0.001 |
| 33 |
266681_at |
AT2G19840
|
[copia-like retrotransposon family, has a 3.5e-301 P-value blast match to GB:CAA31653 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
-0.234 |
0.069 |
| 34 |
249433_at |
AT5G39940
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.232 |
0.019 |
| 35 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-0.225 |
-0.220 |
| 36 |
249392_at |
AT5G40150
|
[Peroxidase superfamily protein] |
-0.222 |
0.089 |
| 37 |
249386_at |
AT5G40060
|
[Disease resistance protein (NBS-LRR class) family] |
-0.217 |
0.056 |
| 38 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.215 |
0.053 |
| 39 |
249499_at |
AT5G39250
|
[F-box family protein] |
-0.213 |
0.056 |
| 40 |
249422_at |
AT5G39760
|
AtHB23, homeobox protein 23, HB23, homeobox protein 23, ZHD10, ZINC FINGER HOMEODOMAIN 10 |
-0.211 |
0.014 |
| 41 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.211 |
-0.061 |
| 42 |
262464_at |
AT1G50280
|
[Phototropic-responsive NPH3 family protein] |
-0.204 |
0.014 |
| 43 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
-0.202 |
-0.244 |
| 44 |
249452_at |
AT5G39500
|
ERMO1, ENDOPLASMIC RETICULUM MORPHOLOGY 1, GNL1, GNOM-like 1 |
-0.201 |
0.024 |
| 45 |
249469_at |
AT5G39320
|
UDG4, UDP-glucose dehydrogenase 4 |
-0.199 |
-0.211 |
| 46 |
249437_at |
AT5G39990
|
GlcAT14A, beta-glucuronosyltransferase 14A |
-0.196 |
-0.058 |
| 47 |
249436_at |
AT5G39980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.193 |
0.124 |