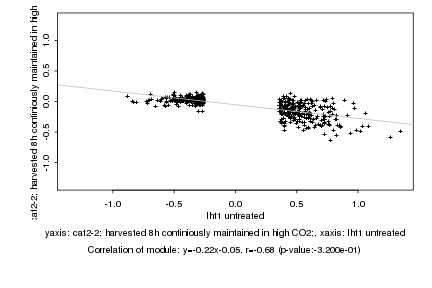
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
lht1 untreated |
Log2 signal ratio
cat2-2; harvested 8h continiously maintained in high CO2; |
| 1 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
1.343 |
-0.483 |
| 2 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
1.261 |
-0.576 |
| 3 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.085 |
-0.401 |
| 4 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
1.053 |
-0.192 |
| 5 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
1.034 |
-0.398 |
| 6 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.016 |
-0.477 |
| 7 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.980 |
-0.471 |
| 8 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.963 |
-0.106 |
| 9 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.958 |
-0.022 |
| 10 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
0.932 |
-0.512 |
| 11 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.912 |
-0.217 |
| 12 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
0.889 |
0.020 |
| 13 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.856 |
-0.406 |
| 14 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.855 |
-0.413 |
| 15 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.848 |
-0.378 |
| 16 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.829 |
-0.390 |
| 17 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.823 |
-0.290 |
| 18 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
0.822 |
-0.221 |
| 19 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.820 |
-0.552 |
| 20 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.812 |
-0.282 |
| 21 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.803 |
-0.128 |
| 22 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.795 |
-0.465 |
| 23 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.793 |
-0.021 |
| 24 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.791 |
0.058 |
| 25 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.786 |
-0.203 |
| 26 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.785 |
-0.105 |
| 27 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
0.772 |
-0.070 |
| 28 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.771 |
-0.633 |
| 29 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.770 |
-0.148 |
| 30 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.765 |
-0.239 |
| 31 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.762 |
-0.375 |
| 32 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.761 |
-0.383 |
| 33 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.758 |
-0.368 |
| 34 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.754 |
-0.179 |
| 35 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.754 |
-0.364 |
| 36 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.752 |
-0.314 |
| 37 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.746 |
-0.087 |
| 38 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
0.736 |
0.035 |
| 39 |
248889_at |
AT5G46230
|
unknown |
0.732 |
-0.241 |
| 40 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
0.731 |
0.007 |
| 41 |
259385_at |
AT1G13470
|
unknown |
0.726 |
-0.334 |
| 42 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
0.725 |
-0.242 |
| 43 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.722 |
-0.358 |
| 44 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.722 |
-0.292 |
| 45 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.720 |
-0.531 |
| 46 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.720 |
-0.292 |
| 47 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
0.719 |
-0.071 |
| 48 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.717 |
-0.282 |
| 49 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.715 |
-0.199 |
| 50 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.713 |
0.022 |
| 51 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
0.713 |
-0.236 |
| 52 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.710 |
-0.389 |
| 53 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.704 |
-0.118 |
| 54 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.700 |
-0.310 |
| 55 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.700 |
-0.399 |
| 56 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.693 |
-0.084 |
| 57 |
258203_at |
AT3G13950
|
unknown |
0.691 |
-0.384 |
| 58 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.686 |
-0.072 |
| 59 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.677 |
-0.366 |
| 60 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.664 |
-0.237 |
| 61 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.656 |
-0.277 |
| 62 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.654 |
0.027 |
| 63 |
255406_at |
AT4G03450
|
[Ankyrin repeat family protein] |
0.650 |
-0.140 |
| 64 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.645 |
-0.387 |
| 65 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.640 |
-0.422 |
| 66 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.638 |
-0.057 |
| 67 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.636 |
-0.282 |
| 68 |
257595_at |
AT3G24750
|
unknown |
0.627 |
0.008 |
| 69 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
0.625 |
-0.328 |
| 70 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.621 |
-0.071 |
| 71 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.617 |
0.021 |
| 72 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.616 |
-0.019 |
| 73 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.616 |
-0.225 |
| 74 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.611 |
-0.320 |
| 75 |
266017_at |
AT2G18690
|
unknown |
0.611 |
-0.272 |
| 76 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.608 |
-0.324 |
| 77 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.605 |
0.045 |
| 78 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
0.605 |
-0.182 |
| 79 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
0.600 |
-0.173 |
| 80 |
258201_at |
AT3G13910
|
unknown |
0.599 |
-0.112 |
| 81 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
0.596 |
-0.214 |
| 82 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.595 |
-0.416 |
| 83 |
261027_at |
AT1G01340
|
ACBK1, ATCNGC10, cyclic nucleotide gated channel 10, CNGC10, cyclic nucleotide gated channel 10 |
0.595 |
-0.278 |
| 84 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.592 |
-0.192 |
| 85 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.591 |
-0.032 |
| 86 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.590 |
-0.250 |
| 87 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.588 |
-0.439 |
| 88 |
267345_at |
AT2G44240
|
unknown |
0.586 |
0.036 |
| 89 |
263854_at |
AT2G04430
|
atnudt5, nudix hydrolase homolog 5, NUDT5, nudix hydrolase homolog 5 |
0.584 |
-0.128 |
| 90 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
0.576 |
-0.272 |
| 91 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.575 |
-0.432 |
| 92 |
258209_at |
AT3G14060
|
unknown |
0.573 |
-0.197 |
| 93 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.571 |
0.028 |
| 94 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.570 |
-0.157 |
| 95 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.568 |
-0.130 |
| 96 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.565 |
-0.090 |
| 97 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.560 |
-0.204 |
| 98 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
0.560 |
-0.334 |
| 99 |
252073_at |
AT3G51750
|
unknown |
0.557 |
-0.022 |
| 100 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.557 |
-0.388 |
| 101 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.556 |
-0.250 |
| 102 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
0.555 |
-0.471 |
| 103 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
0.553 |
-0.054 |
| 104 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.545 |
-0.328 |
| 105 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.544 |
-0.185 |
| 106 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
0.543 |
-0.080 |
| 107 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.538 |
0.022 |
| 108 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.537 |
-0.242 |
| 109 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.532 |
-0.002 |
| 110 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
0.529 |
-0.060 |
| 111 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.528 |
-0.099 |
| 112 |
266464_at |
AT2G47800
|
ABCC4, ATP-binding cassette C4, ATMRP4, multidrug resistance-associated protein 4, EST3, MRP4, multidrug resistance-associated protein 4 |
0.527 |
-0.215 |
| 113 |
258182_at |
AT3G21500
|
DXL1, DXS-like 1, DXPS1, 1-deoxy-D-xylulose 5-phosphate synthase 1, DXS2, 1-deoxy-D-xylulose 5-phosphate (DXP) synthase 2 |
0.525 |
0.030 |
| 114 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.522 |
-0.287 |
| 115 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.522 |
-0.202 |
| 116 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.521 |
-0.324 |
| 117 |
245401_at |
AT4G17670
|
unknown |
0.518 |
-0.155 |
| 118 |
247779_at |
AT5G58760
|
DDB2, damaged DNA binding 2 |
0.518 |
-0.019 |
| 119 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
0.517 |
-0.244 |
| 120 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.516 |
-0.110 |
| 121 |
261866_at |
AT1G50420
|
SCL-3, SCARECROW-LIKE 3, SCL3, scarecrow-like 3 |
0.516 |
-0.083 |
| 122 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.514 |
-0.183 |
| 123 |
249454_at |
AT5G39520
|
unknown |
0.514 |
-0.411 |
| 124 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.510 |
-0.030 |
| 125 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.510 |
-0.052 |
| 126 |
251633_at |
AT3G57460
|
[catalytics] |
0.509 |
-0.205 |
| 127 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.508 |
-0.150 |
| 128 |
265025_at |
AT1G24575
|
unknown |
0.507 |
-0.081 |
| 129 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.504 |
-0.320 |
| 130 |
256883_at |
AT3G26440
|
unknown |
0.503 |
-0.208 |
| 131 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.503 |
-0.152 |
| 132 |
259856_at |
AT1G68440
|
unknown |
0.502 |
-0.015 |
| 133 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.491 |
-0.181 |
| 134 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.490 |
-0.356 |
| 135 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
0.488 |
-0.203 |
| 136 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.487 |
-0.282 |
| 137 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.485 |
-0.207 |
| 138 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
0.483 |
-0.071 |
| 139 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.482 |
-0.178 |
| 140 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
0.482 |
-0.175 |
| 141 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.481 |
0.084 |
| 142 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.478 |
-0.100 |
| 143 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.474 |
-0.186 |
| 144 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.474 |
-0.257 |
| 145 |
246125_at |
AT5G19875
|
unknown |
0.474 |
-0.084 |
| 146 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.472 |
-0.025 |
| 147 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.470 |
-0.275 |
| 148 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
0.470 |
-0.088 |
| 149 |
251020_at |
AT5G02270
|
ABCI20, ATP-binding cassette I20, ATNAP9, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 9, NAP9, non-intrinsic ABC protein 9 |
0.469 |
0.021 |
| 150 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
0.468 |
-0.299 |
| 151 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.467 |
-0.079 |
| 152 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.467 |
-0.120 |
| 153 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
0.467 |
-0.088 |
| 154 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.467 |
-0.021 |
| 155 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.466 |
-0.199 |
| 156 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.463 |
-0.047 |
| 157 |
247740_at |
AT5G58940
|
CRCK1, calmodulin-binding receptor-like cytoplasmic kinase 1 |
0.463 |
-0.109 |
| 158 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.461 |
0.030 |
| 159 |
266376_at |
AT2G14620
|
XTH10, xyloglucan endotransglucosylase/hydrolase 10 |
0.460 |
-0.131 |
| 160 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
0.459 |
-0.043 |
| 161 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.459 |
-0.165 |
| 162 |
249377_at |
AT5G40690
|
unknown |
0.452 |
-0.243 |
| 163 |
259424_at |
AT1G13830
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.451 |
-0.085 |
| 164 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.451 |
-0.114 |
| 165 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.449 |
0.036 |
| 166 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.448 |
-0.105 |
| 167 |
251422_at |
AT3G60540
|
[Preprotein translocase Sec, Sec61-beta subunit protein] |
0.448 |
-0.125 |
| 168 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
0.445 |
-0.332 |
| 169 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.445 |
-0.277 |
| 170 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.445 |
-0.266 |
| 171 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.444 |
-0.174 |
| 172 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.442 |
0.147 |
| 173 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.440 |
-0.208 |
| 174 |
264752_at |
AT1G23010
|
LPR1, Low Phosphate Root1 |
0.437 |
-0.019 |
| 175 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.437 |
0.035 |
| 176 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.436 |
-0.234 |
| 177 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.435 |
0.000 |
| 178 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
0.435 |
0.027 |
| 179 |
260396_at |
AT1G69720
|
HO3, heme oxygenase 3 |
0.434 |
-0.148 |
| 180 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.432 |
-0.035 |
| 181 |
260648_at |
AT1G08050
|
[Zinc finger (C3HC4-type RING finger) family protein] |
0.432 |
-0.232 |
| 182 |
256627_at |
AT3G19970
|
[alpha/beta-Hydrolases superfamily protein] |
0.431 |
0.052 |
| 183 |
265943_at |
AT2G19570
|
AT-CDA1, CDA1, cytidine deaminase 1, DESZ |
0.429 |
-0.057 |
| 184 |
255319_at |
AT4G04220
|
AtRLP46, receptor like protein 46, RLP46, receptor like protein 46 |
0.426 |
-0.213 |
| 185 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
0.426 |
-0.166 |
| 186 |
257314_at |
AT3G26590
|
[MATE efflux family protein] |
0.423 |
-0.114 |
| 187 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
0.423 |
-0.119 |
| 188 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.421 |
-0.165 |
| 189 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
0.420 |
0.012 |
| 190 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
0.412 |
-0.068 |
| 191 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.410 |
-0.214 |
| 192 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
0.410 |
0.084 |
| 193 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.408 |
-0.005 |
| 194 |
250994_at |
AT5G02490
|
AtHsp70-2, Hsp70-2 |
0.408 |
-0.090 |
| 195 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
0.407 |
0.002 |
| 196 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.405 |
-0.198 |
| 197 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.404 |
-0.065 |
| 198 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.402 |
-0.318 |
| 199 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
0.402 |
-0.049 |
| 200 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
0.400 |
-0.165 |
| 201 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
0.399 |
-0.094 |
| 202 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.399 |
0.051 |
| 203 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.399 |
-0.466 |
| 204 |
254253_at |
AT4G23320
|
CRK24, cysteine-rich RLK (RECEPTOR-like protein kinase) 24 |
0.398 |
-0.193 |
| 205 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.396 |
-0.055 |
| 206 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.394 |
-0.348 |
| 207 |
259489_at |
AT1G15790
|
unknown |
0.394 |
-0.128 |
| 208 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.394 |
-0.405 |
| 209 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.392 |
-0.377 |
| 210 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.391 |
-0.273 |
| 211 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.387 |
-0.094 |
| 212 |
253401_at |
AT4G32870
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.387 |
-0.276 |
| 213 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
0.385 |
0.065 |
| 214 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.385 |
0.093 |
| 215 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.382 |
-0.335 |
| 216 |
252611_at |
AT3G45130
|
LAS1, lanosterol synthase 1 |
0.379 |
-0.055 |
| 217 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
0.379 |
-0.202 |
| 218 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.376 |
-0.075 |
| 219 |
248205_at |
AT5G54300
|
unknown |
0.375 |
-0.012 |
| 220 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.374 |
-0.145 |
| 221 |
267178_at |
AT2G37750
|
unknown |
0.372 |
-0.329 |
| 222 |
266596_at |
AT2G46150
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.371 |
-0.154 |
| 223 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
0.371 |
0.002 |
| 224 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.371 |
-0.061 |
| 225 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.370 |
0.033 |
| 226 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.370 |
-0.204 |
| 227 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.369 |
-0.355 |
| 228 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
0.368 |
-0.137 |
| 229 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.368 |
-0.293 |
| 230 |
245566_at |
AT4G14610
|
[pseudogene] |
0.368 |
-0.184 |
| 231 |
255627_at |
AT4G00955
|
unknown |
0.364 |
-0.121 |
| 232 |
262408_at |
AT1G34750
|
[Protein phosphatase 2C family protein] |
0.364 |
-0.060 |
| 233 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.363 |
-0.101 |
| 234 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.361 |
-0.319 |
| 235 |
253101_at |
AT4G37430
|
CYP81F1, CYTOCHROME P450 MONOOXYGENASE 81F1, CYP91A2, cytochrome P450, family 91, subfamily A, polypeptide 2 |
0.361 |
-0.016 |
| 236 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.360 |
-0.158 |
| 237 |
252421_at |
AT3G47540
|
[Chitinase family protein] |
0.359 |
-0.106 |
| 238 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
0.359 |
-0.058 |
| 239 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.358 |
-0.163 |
| 240 |
264692_at |
AT1G70000
|
[myb-like transcription factor family protein] |
0.358 |
-0.052 |
| 241 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
0.355 |
-0.016 |
| 242 |
251739_at |
AT3G56170
|
CAN, Ca-2+ dependent nuclease, CAN1, calcium dependent nuclease 1 |
0.355 |
-0.065 |
| 243 |
256529_at |
AT1G33260
|
[Protein kinase superfamily protein] |
0.355 |
0.044 |
| 244 |
255259_at |
AT4G05020
|
NDB2, NAD(P)H dehydrogenase B2 |
0.354 |
-0.061 |
| 245 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.882 |
0.096 |
| 246 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.845 |
0.007 |
| 247 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.837 |
-0.016 |
| 248 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.810 |
-0.010 |
| 249 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.733 |
0.010 |
| 250 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.723 |
-0.025 |
| 251 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.715 |
0.017 |
| 252 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.703 |
0.022 |
| 253 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.700 |
0.119 |
| 254 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.691 |
0.038 |
| 255 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
-0.652 |
-0.078 |
| 256 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.643 |
0.024 |
| 257 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.612 |
0.006 |
| 258 |
261248_at |
AT1G20030
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.609 |
0.035 |
| 259 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.608 |
0.048 |
| 260 |
250327_at |
AT5G12050
|
unknown |
-0.602 |
0.059 |
| 261 |
258225_at |
AT3G15630
|
unknown |
-0.583 |
-0.056 |
| 262 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.573 |
0.068 |
| 263 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.573 |
-0.070 |
| 264 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-0.569 |
-0.004 |
| 265 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
-0.565 |
0.051 |
| 266 |
248509_at |
AT5G50335
|
unknown |
-0.563 |
0.016 |
| 267 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.562 |
0.067 |
| 268 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.548 |
-0.049 |
| 269 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.542 |
0.075 |
| 270 |
264314_at |
AT1G70420
|
unknown |
-0.537 |
0.030 |
| 271 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.534 |
0.003 |
| 272 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.521 |
0.024 |
| 273 |
252464_at |
AT3G47160
|
[RING/U-box superfamily protein] |
-0.519 |
0.009 |
| 274 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.518 |
0.035 |
| 275 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.507 |
0.129 |
| 276 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.507 |
0.109 |
| 277 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-0.503 |
0.060 |
| 278 |
250412_at |
AT5G11150
|
ATVAMP713, vesicle-associated membrane protein 713, VAMP713, VAMP713, vesicle-associated membrane protein 713 |
-0.502 |
0.047 |
| 279 |
247474_at |
AT5G62280
|
unknown |
-0.502 |
0.158 |
| 280 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.501 |
0.109 |
| 281 |
262236_at |
AT1G48330
|
unknown |
-0.498 |
0.066 |
| 282 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-0.496 |
0.082 |
| 283 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.489 |
0.012 |
| 284 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.487 |
-0.016 |
| 285 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.482 |
-0.033 |
| 286 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
-0.479 |
0.032 |
| 287 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
-0.478 |
0.014 |
| 288 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
-0.478 |
0.036 |
| 289 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.477 |
0.015 |
| 290 |
265481_at |
AT2G15960
|
unknown |
-0.475 |
-0.015 |
| 291 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.470 |
0.009 |
| 292 |
246929_at |
AT5G25210
|
unknown |
-0.467 |
-0.079 |
| 293 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
-0.466 |
0.045 |
| 294 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
-0.463 |
0.024 |
| 295 |
254193_at |
AT4G23870
|
unknown |
-0.459 |
0.004 |
| 296 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.454 |
-0.016 |
| 297 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
-0.453 |
0.068 |
| 298 |
245602_at |
AT4G14270
|
unknown |
-0.453 |
-0.000 |
| 299 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.452 |
0.101 |
| 300 |
250127_at |
AT5G16380
|
unknown |
-0.451 |
0.009 |
| 301 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
-0.450 |
0.063 |
| 302 |
253317_at |
AT4G33960
|
unknown |
-0.444 |
0.019 |
| 303 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
-0.443 |
0.033 |
| 304 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.443 |
0.035 |
| 305 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.440 |
0.054 |
| 306 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.439 |
0.001 |
| 307 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.433 |
0.074 |
| 308 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.432 |
0.032 |
| 309 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.429 |
0.030 |
| 310 |
260411_at |
AT1G69890
|
unknown |
-0.428 |
0.011 |
| 311 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
-0.426 |
0.014 |
| 312 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.423 |
0.035 |
| 313 |
262170_at |
AT1G74940
|
unknown |
-0.412 |
0.016 |
| 314 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.410 |
0.047 |
| 315 |
258939_at |
AT3G10020
|
unknown |
-0.406 |
-0.025 |
| 316 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
-0.405 |
0.010 |
| 317 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.405 |
0.099 |
| 318 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.404 |
0.023 |
| 319 |
258367_at |
AT3G14370
|
WAG2 |
-0.403 |
0.084 |
| 320 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.401 |
0.036 |
| 321 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
-0.401 |
0.050 |
| 322 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.397 |
-0.013 |
| 323 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.395 |
0.013 |
| 324 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
-0.394 |
0.025 |
| 325 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.393 |
-0.001 |
| 326 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.392 |
0.092 |
| 327 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
-0.392 |
0.029 |
| 328 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.391 |
0.091 |
| 329 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.388 |
0.107 |
| 330 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.384 |
0.079 |
| 331 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
-0.381 |
0.022 |
| 332 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.380 |
0.072 |
| 333 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
-0.379 |
-0.061 |
| 334 |
267077_at |
AT2G40970
|
MYBC1 |
-0.378 |
0.019 |
| 335 |
250751_at |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.377 |
0.128 |
| 336 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
-0.376 |
0.072 |
| 337 |
254645_at |
AT4G18520
|
SEL1, SEEDLING LETHAL 1 |
-0.373 |
0.025 |
| 338 |
255943_at |
AT1G22370
|
AtUGT85A5, UDP-glucosyl transferase 85A5, UGT85A5, UDP-glucosyl transferase 85A5 |
-0.372 |
0.071 |
| 339 |
250900_at |
AT5G03470
|
Protein phosphatase 2A regulatory B subunit family protein |
-0.370 |
0.030 |
| 340 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
-0.368 |
0.126 |
| 341 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.368 |
0.065 |
| 342 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.367 |
0.071 |
| 343 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.363 |
0.045 |
| 344 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
-0.363 |
0.025 |
| 345 |
253981_at |
AT4G26670
|
[Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein] |
-0.362 |
0.029 |
| 346 |
247903_at |
AT5G57340
|
unknown |
-0.362 |
-0.011 |
| 347 |
248502_at |
AT5G50450
|
[HCP-like superfamily protein with MYND-type zinc finger] |
-0.357 |
0.039 |
| 348 |
247585_at |
AT5G60680
|
unknown |
-0.357 |
0.051 |
| 349 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.354 |
0.097 |
| 350 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.354 |
0.018 |
| 351 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
-0.351 |
-0.058 |
| 352 |
254089_at |
AT4G24800
|
ECIP1, EIN2 C-terminus Interacting Protein 1 |
-0.351 |
0.030 |
| 353 |
254424_at |
AT4G21510
|
AtFBS2, FBS2, F-box stress induced 2 |
-0.346 |
0.031 |
| 354 |
264900_at |
AT1G23080
|
ATPIN7, ARABIDOPSIS PIN-FORMED 7, PIN7, PIN-FORMED 7 |
-0.345 |
0.062 |
| 355 |
264653_at |
AT1G08980
|
AMI1, amidase 1, ATAMI1, AMIDASE-LIKE PROTEIN 1, ATTOC64-I, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-I, TOC64-I, TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-I |
-0.344 |
0.022 |
| 356 |
261203_at |
AT1G12845
|
unknown |
-0.343 |
0.028 |
| 357 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.341 |
0.045 |
| 358 |
253581_at |
AT4G30660
|
[Low temperature and salt responsive protein family] |
-0.341 |
0.021 |
| 359 |
265539_at |
AT2G15830
|
unknown |
-0.340 |
-0.025 |
| 360 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.339 |
0.064 |
| 361 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.336 |
0.023 |
| 362 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.336 |
0.041 |
| 363 |
259080_at |
AT3G04910
|
ATWNK1, WNK1, with no lysine (K) kinase 1, ZIK4 |
-0.334 |
0.031 |
| 364 |
262114_at |
AT1G02860
|
BAH1, BENZOIC ACID HYPERSENSITIVE 1, NLA, nitrogen limitation adaptation |
-0.334 |
0.029 |
| 365 |
267280_at |
AT2G19450
|
ABX45, AS11, ATDGAT, Arabidopsis thaliana acyl-CoA:diacylglycerol acyltransferase, AtDGAT1, DGAT1, acyl-CoA:diacylglycerol acyltransferase 1, RDS1, TAG1, TRIACYLGLYCEROL BIOSYNTHESIS DEFECT 1 |
-0.333 |
0.052 |
| 366 |
247443_at |
AT5G62720
|
[Integral membrane HPP family protein] |
-0.332 |
0.035 |
| 367 |
259163_at |
AT3G01490
|
[Protein kinase superfamily protein] |
-0.331 |
0.044 |
| 368 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.329 |
0.045 |
| 369 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.329 |
0.031 |
| 370 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.328 |
0.037 |
| 371 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
-0.328 |
0.008 |
| 372 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.328 |
0.030 |
| 373 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.327 |
-0.074 |
| 374 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.325 |
-0.047 |
| 375 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.324 |
0.089 |
| 376 |
262892_at |
AT1G79440
|
ALDH5F1, aldehyde dehydrogenase 5F1, ENF1, ENLARGED FIL EXPRESSION DOMAIN 1, SSADH, SUCCINIC SEMIALDEHYDE DEHYDROGENASE, SSADH1, SUCCINIC SEMIALDEHYDE DEHYDROGENASE 1 |
-0.324 |
0.047 |
| 377 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
-0.323 |
0.156 |
| 378 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.322 |
0.075 |
| 379 |
253493_at |
AT4G31820
|
ENP, ENHANCER OF PINOID, MAB4, MACCHI-BOU 4, NPY1, NAKED PINS IN YUC MUTANTS 1 |
-0.321 |
0.083 |
| 380 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
-0.320 |
0.032 |
| 381 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.319 |
0.023 |
| 382 |
253799_at |
AT4G28140
|
[Integrase-type DNA-binding superfamily protein] |
-0.319 |
0.007 |
| 383 |
245761_at |
AT1G66890
|
unknown |
-0.319 |
0.010 |
| 384 |
252036_at |
AT3G52070
|
unknown |
-0.317 |
-0.006 |
| 385 |
255255_at |
AT4G05070
|
[Wound-responsive family protein] |
-0.316 |
-0.041 |
| 386 |
252956_at |
AT4G38580
|
ATFP6, farnesylated protein 6, FP6, farnesylated protein 6, HIPP26, HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN 26 |
-0.316 |
0.002 |
| 387 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.316 |
0.048 |
| 388 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
-0.315 |
0.039 |
| 389 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.314 |
0.057 |
| 390 |
260034_at |
AT1G68810
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.313 |
0.072 |
| 391 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.312 |
-0.009 |
| 392 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.311 |
-0.010 |
| 393 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.311 |
0.101 |
| 394 |
254919_at |
AT4G11360
|
RHA1B, RING-H2 finger A1B |
-0.311 |
0.052 |
| 395 |
266225_at |
AT2G28900
|
ATOEP16-1, outer plastid envelope protein 16-1, ATOEP16-L, OUTER PLASTID ENVELOPE PROTEIN 16-L, OEP16, outer envelope protein 16, OEP16-1, outer plastid envelope protein 16-1 |
-0.310 |
0.015 |
| 396 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
-0.310 |
0.046 |
| 397 |
250158_at |
AT5G15190
|
unknown |
-0.310 |
0.010 |
| 398 |
263002_at |
AT1G54200
|
unknown |
-0.307 |
0.103 |
| 399 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.307 |
0.067 |
| 400 |
247043_at |
AT5G66880
|
SNRK2-3, SUCROSE NONFERMENTING 1 (SNF1)-RELATED PROTEIN KINASE 2-3, SNRK2.3, sucrose nonfermenting 1(SNF1)-related protein kinase 2.3, SRK2I |
-0.306 |
0.013 |
| 401 |
255899_at |
AT1G17970
|
[RING/U-box superfamily protein] |
-0.305 |
0.070 |
| 402 |
246829_at |
AT5G26570
|
ATGWD3, OK1, PWD, PHOSPHOGLUCAN WATER DIKINASE |
-0.305 |
0.021 |
| 403 |
266929_at |
AT2G45850
|
AHL9, AT-hook motif nuclear localized protein 9 |
-0.304 |
0.040 |
| 404 |
261026_at |
AT1G01240
|
unknown |
-0.304 |
0.029 |
| 405 |
266259_at |
AT2G27830
|
unknown |
-0.303 |
-0.047 |
| 406 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.303 |
-0.150 |
| 407 |
258833_at |
AT3G07274
|
|
-0.303 |
0.072 |
| 408 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
-0.302 |
0.018 |
| 409 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
-0.302 |
0.046 |
| 410 |
251225_at |
AT3G62660
|
GATL7, galacturonosyltransferase-like 7 |
-0.302 |
0.048 |
| 411 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-0.301 |
-0.040 |
| 412 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.300 |
0.045 |
| 413 |
249456_at |
AT5G39410
|
[Saccharopine dehydrogenase ] |
-0.300 |
0.034 |
| 414 |
255232_at |
AT4G05330
|
AGD13, ARF-GAP domain 13 |
-0.300 |
0.048 |
| 415 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.299 |
0.085 |
| 416 |
261187_at |
AT1G32860
|
[Glycosyl hydrolase superfamily protein] |
-0.298 |
0.093 |
| 417 |
255417_at |
AT4G03190
|
AFB1, AUXIN SIGNALING F BOX PROTEIN 1, ATGRH1, GRH1, GRR1-like protein 1 |
-0.297 |
0.061 |
| 418 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.297 |
-0.052 |
| 419 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
-0.297 |
0.094 |
| 420 |
259996_at |
AT1G67910
|
unknown |
-0.296 |
0.025 |
| 421 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
-0.294 |
-0.020 |
| 422 |
261366_at |
AT1G53100
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.293 |
-0.034 |
| 423 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.292 |
0.079 |
| 424 |
262881_at |
AT1G64890
|
[Major facilitator superfamily protein] |
-0.291 |
0.040 |
| 425 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.291 |
0.096 |
| 426 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
-0.291 |
-0.030 |
| 427 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.290 |
0.037 |
| 428 |
261230_at |
AT1G20010
|
TUB5, tubulin beta-5 chain |
-0.290 |
0.024 |
| 429 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
-0.289 |
0.043 |
| 430 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.289 |
0.046 |
| 431 |
266803_at |
AT2G28930
|
APK1B, protein kinase 1B, PK1B, protein kinase 1B |
-0.288 |
0.040 |
| 432 |
249220_at |
AT5G42420
|
[Nucleotide-sugar transporter family protein] |
-0.288 |
0.036 |
| 433 |
247151_at |
AT5G65640
|
bHLH093, beta HLH protein 93 |
-0.287 |
-0.005 |
| 434 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
-0.286 |
0.078 |
| 435 |
260671_at |
AT1G19310
|
[RING/U-box superfamily protein] |
-0.286 |
0.016 |
| 436 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
-0.285 |
-0.060 |
| 437 |
258374_at |
AT3G14360
|
[alpha/beta-Hydrolases superfamily protein] |
-0.284 |
0.100 |
| 438 |
258213_at |
AT3G17950
|
unknown |
-0.284 |
0.020 |
| 439 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.284 |
0.048 |
| 440 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.283 |
0.048 |
| 441 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
-0.283 |
0.026 |
| 442 |
258196_at |
AT3G13980
|
unknown |
-0.282 |
0.111 |
| 443 |
249128_at |
AT5G43440
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.281 |
-0.001 |
| 444 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
-0.281 |
0.056 |
| 445 |
264799_at |
AT1G08550
|
AVDE1, ARABIDOPSIS VIOLAXANTHIN DE-EPOXIDASE 1, NPQ1, non-photochemical quenching 1 |
-0.281 |
0.032 |
| 446 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.281 |
0.131 |
| 447 |
256220_at |
AT1G56230
|
unknown |
-0.281 |
0.026 |
| 448 |
252184_at |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
-0.280 |
0.039 |
| 449 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.278 |
0.049 |
| 450 |
250524_at |
AT5G08520
|
[Duplicated homeodomain-like superfamily protein] |
-0.278 |
0.012 |
| 451 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.277 |
0.028 |
| 452 |
266016_at |
AT2G18670
|
[RING/U-box superfamily protein] |
-0.277 |
0.030 |
| 453 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
-0.276 |
0.008 |
| 454 |
247278_at |
AT5G64380
|
[Inositol monophosphatase family protein] |
-0.276 |
0.048 |
| 455 |
255726_at |
AT1G25530
|
[Transmembrane amino acid transporter family protein] |
-0.276 |
0.088 |
| 456 |
267569_at |
AT2G30790
|
PSBP-2, photosystem II subunit P-2 |
-0.275 |
0.044 |
| 457 |
252360_at |
AT3G48480
|
[Cysteine proteinases superfamily protein] |
-0.275 |
-0.034 |
| 458 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
-0.274 |
-0.148 |
| 459 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.274 |
0.069 |
| 460 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-0.274 |
0.089 |
| 461 |
253812_at |
AT4G28240
|
[Wound-responsive family protein] |
-0.274 |
-0.001 |
| 462 |
262478_at |
AT1G11170
|
unknown |
-0.272 |
0.021 |
| 463 |
262296_at |
AT1G27630
|
CYCT1;3, cyclin T 1;3 |
-0.272 |
0.066 |
| 464 |
249069_at |
AT5G44010
|
unknown |
-0.272 |
0.056 |
| 465 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
-0.271 |
-0.062 |
| 466 |
266707_at |
AT2G03310
|
unknown |
-0.271 |
0.015 |
| 467 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
-0.271 |
0.028 |
| 468 |
249742_at |
AT5G24490
|
[30S ribosomal protein, putative] |
-0.270 |
0.012 |
| 469 |
263912_at |
AT2G36390
|
BE3, BRANCHING ENZYME 3, SBE2.1, starch branching enzyme 2.1 |
-0.270 |
0.033 |
| 470 |
251036_at |
AT5G02160
|
unknown |
-0.270 |
0.015 |
| 471 |
264379_at |
AT2G25200
|
unknown |
-0.269 |
0.010 |
| 472 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.269 |
0.133 |
| 473 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.268 |
0.077 |
| 474 |
246253_at |
AT4G37260
|
ATMYB73, MYB73, myb domain protein 73 |
-0.268 |
0.037 |
| 475 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
-0.268 |
-0.005 |
| 476 |
251235_at |
AT3G62860
|
[alpha/beta-Hydrolases superfamily protein] |
-0.267 |
0.063 |
| 477 |
246881_at |
AT5G26040
|
HDA2, histone deacetylase 2 |
-0.267 |
0.118 |
| 478 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.267 |
0.031 |
| 479 |
247191_at |
AT5G65310
|
ATHB-5, ATHB5, homeobox protein 5, HB5, homeobox protein 5 |
-0.267 |
0.039 |
| 480 |
262920_at |
AT1G79500
|
AtkdsA1 |
-0.266 |
0.019 |
| 481 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
-0.265 |
0.032 |
| 482 |
254011_at |
AT4G26370
|
[antitermination NusB domain-containing protein] |
-0.265 |
0.043 |
| 483 |
264854_at |
AT2G17450
|
RHA3A, RING-H2 finger A3A |
-0.265 |
0.019 |
| 484 |
266119_at |
AT2G02100
|
LCR69, low-molecular-weight cysteine-rich 69, PDF2.2 |
-0.265 |
-0.005 |
| 485 |
254659_at |
AT4G18240
|
ATSS4, ARABIDOPSIS THALIANA STARCH SYNTHASE 4, SS4, starch synthase 4, SSIV, STARCH SYNTHASE 4 |
-0.265 |
0.016 |
| 486 |
246942_at |
AT5G25430
|
[HCO3- transporter family] |
-0.264 |
0.015 |
| 487 |
266988_at |
AT2G39310
|
JAL22, jacalin-related lectin 22 |
-0.264 |
-0.017 |