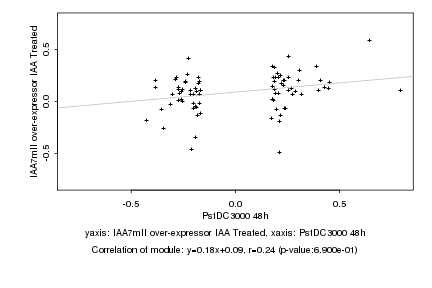
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
PstDC3000 48h |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
267345_at |
AT2G44240
|
unknown |
0.790 |
0.107 |
| 2 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.643 |
0.594 |
| 3 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
0.451 |
0.186 |
| 4 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.445 |
0.127 |
| 5 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.425 |
0.138 |
| 6 |
259385_at |
AT1G13470
|
unknown |
0.406 |
0.206 |
| 7 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
0.397 |
0.108 |
| 8 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.387 |
0.346 |
| 9 |
252346_at |
AT3G48650
|
[pseudogene] |
0.316 |
0.074 |
| 10 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.303 |
0.303 |
| 11 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.299 |
0.211 |
| 12 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
0.285 |
0.101 |
| 13 |
249940_at |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
0.269 |
0.074 |
| 14 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
0.265 |
0.131 |
| 15 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.254 |
0.241 |
| 16 |
245566_at |
AT4G14610
|
[pseudogene] |
0.254 |
0.108 |
| 17 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
0.254 |
0.442 |
| 18 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
0.237 |
-0.059 |
| 19 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.235 |
-0.066 |
| 20 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.234 |
0.211 |
| 21 |
252345_at |
AT3G48640
|
unknown |
0.227 |
0.204 |
| 22 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.226 |
0.160 |
| 23 |
249096_at |
AT5G43910
|
[pfkB-like carbohydrate kinase family protein] |
0.219 |
0.183 |
| 24 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.215 |
-0.129 |
| 25 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.213 |
0.257 |
| 26 |
254901_at |
AT4G11530
|
CRK34, cysteine-rich RLK (RECEPTOR-like protein kinase) 34 |
0.208 |
-0.489 |
| 27 |
263068_at |
AT2G17580
|
[Polynucleotide adenylyltransferase family protein] |
0.207 |
-0.189 |
| 28 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.205 |
0.233 |
| 29 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
0.202 |
0.079 |
| 30 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.197 |
0.277 |
| 31 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.194 |
-0.068 |
| 32 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.192 |
0.086 |
| 33 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.192 |
0.240 |
| 34 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.187 |
0.200 |
| 35 |
246236_at |
AT4G36470
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.186 |
0.117 |
| 36 |
251232_at |
AT3G62780
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.183 |
0.337 |
| 37 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
0.181 |
0.234 |
| 38 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
0.178 |
0.015 |
| 39 |
250679_at |
AT5G06550
|
JMJ22, Jumonji domain-containing protein 22 |
0.177 |
0.025 |
| 40 |
262802_at |
AT1G20930
|
CDKB2;2, cyclin-dependent kinase B2;2 |
0.176 |
0.022 |
| 41 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
0.174 |
0.145 |
| 42 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.173 |
0.340 |
| 43 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.172 |
-0.161 |
| 44 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.429 |
-0.176 |
| 45 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.388 |
0.203 |
| 46 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.386 |
0.140 |
| 47 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
-0.359 |
-0.076 |
| 48 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.348 |
-0.253 |
| 49 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
-0.313 |
-0.028 |
| 50 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
-0.303 |
0.076 |
| 51 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
-0.289 |
0.216 |
| 52 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.285 |
0.235 |
| 53 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
-0.277 |
0.010 |
| 54 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.277 |
0.142 |
| 55 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.274 |
0.117 |
| 56 |
256603_at |
AT3G28270
|
unknown |
-0.270 |
0.084 |
| 57 |
247764_at |
AT5G59190
|
[subtilase family protein] |
-0.259 |
0.029 |
| 58 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
-0.259 |
0.099 |
| 59 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
-0.258 |
0.004 |
| 60 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.256 |
0.123 |
| 61 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
-0.242 |
0.192 |
| 62 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-0.240 |
0.193 |
| 63 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
-0.233 |
0.266 |
| 64 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
-0.229 |
0.420 |
| 65 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
-0.217 |
0.075 |
| 66 |
256924_at |
AT3G29590
|
AT5MAT |
-0.216 |
0.112 |
| 67 |
258367_at |
AT3G14370
|
WAG2 |
-0.211 |
-0.453 |
| 68 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.206 |
0.072 |
| 69 |
261437_at |
AT1G07745
|
ATRAD51D, RAD51D, homolog of RAD51 D, SSN1, SUPPRESOR OF SNI1 |
-0.204 |
-0.062 |
| 70 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.204 |
-0.010 |
| 71 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
-0.195 |
0.128 |
| 72 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.194 |
-0.344 |
| 73 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
-0.194 |
-0.040 |
| 74 |
258225_at |
AT3G15630
|
unknown |
-0.190 |
0.097 |
| 75 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
-0.187 |
-0.054 |
| 76 |
257798_at |
AT3G15950
|
NAI2 |
-0.183 |
-0.131 |
| 77 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.180 |
0.174 |
| 78 |
256097_at |
AT1G13670
|
unknown |
-0.179 |
0.235 |
| 79 |
245572_at |
AT4G14720
|
PPD2, PEAPOD 2, TIFY4B |
-0.176 |
-0.010 |
| 80 |
266823_at |
AT2G44930
|
unknown |
-0.175 |
0.077 |
| 81 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
-0.175 |
0.069 |
| 82 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
-0.174 |
0.198 |
| 83 |
266977_at |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.170 |
0.113 |
| 84 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.169 |
0.108 |
| 85 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.168 |
-0.109 |