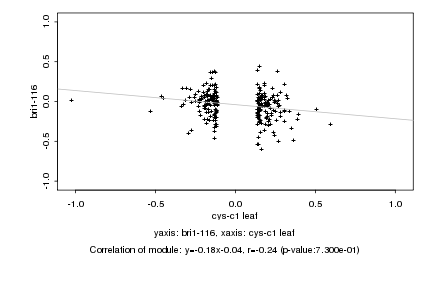
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
cys-c1 leaf |
Log2 signal ratio
bri1-116 |
| 1 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
0.593 |
-0.287 |
| 2 |
262373_at |
AT1G73120
|
unknown |
0.501 |
-0.096 |
| 3 |
263182_at |
AT1G05575
|
unknown |
0.390 |
-0.157 |
| 4 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
0.387 |
-0.215 |
| 5 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.359 |
-0.482 |
| 6 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.348 |
-0.332 |
| 7 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.335 |
-0.117 |
| 8 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.321 |
0.038 |
| 9 |
258653_at |
AT3G09870
|
SAUR48, SMALL AUXIN UPREGULATED RNA 48 |
0.318 |
0.080 |
| 10 |
253044_at |
AT4G37290
|
unknown |
0.306 |
-0.242 |
| 11 |
245692_at |
AT5G04150
|
BHLH101 |
0.305 |
0.221 |
| 12 |
245755_at |
AT1G35210
|
unknown |
0.303 |
-0.125 |
| 13 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.301 |
-0.110 |
| 14 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.281 |
0.121 |
| 15 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.280 |
-0.177 |
| 16 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.278 |
-0.133 |
| 17 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.274 |
-0.047 |
| 18 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
0.267 |
-0.055 |
| 19 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.267 |
0.025 |
| 20 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.265 |
-0.490 |
| 21 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.263 |
-0.020 |
| 22 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
0.262 |
0.389 |
| 23 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
0.261 |
0.076 |
| 24 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.252 |
-0.230 |
| 25 |
267623_at |
AT2G39650
|
unknown |
0.248 |
-0.116 |
| 26 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.246 |
0.005 |
| 27 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
0.244 |
-0.001 |
| 28 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.243 |
-0.421 |
| 29 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
0.240 |
0.016 |
| 30 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.238 |
-0.101 |
| 31 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
0.237 |
-0.385 |
| 32 |
252347_at |
AT3G48130
|
RSU1 |
0.235 |
0.084 |
| 33 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
0.231 |
0.056 |
| 34 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
0.228 |
0.174 |
| 35 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
0.227 |
0.058 |
| 36 |
245041_at |
AT2G26530
|
AR781 |
0.222 |
-0.142 |
| 37 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.221 |
-0.168 |
| 38 |
256096_at |
AT1G13650
|
unknown |
0.219 |
0.029 |
| 39 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
0.218 |
-0.286 |
| 40 |
251531_at |
AT3G58550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.214 |
-0.076 |
| 41 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.208 |
-0.225 |
| 42 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.207 |
-0.030 |
| 43 |
247047_at |
AT5G66650
|
unknown |
0.205 |
-0.294 |
| 44 |
246001_at |
AT5G20790
|
unknown |
0.204 |
-0.036 |
| 45 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
0.203 |
-0.244 |
| 46 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
0.201 |
0.014 |
| 47 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.199 |
-0.181 |
| 48 |
264573_at |
AT1G05310
|
[Pectin lyase-like superfamily protein] |
0.199 |
-0.062 |
| 49 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.197 |
-0.006 |
| 50 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.195 |
-0.150 |
| 51 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.194 |
-0.035 |
| 52 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.194 |
-0.022 |
| 53 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
0.193 |
-0.194 |
| 54 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.191 |
-0.213 |
| 55 |
246796_at |
AT5G26770
|
unknown |
0.187 |
-0.038 |
| 56 |
245697_at |
AT5G04200
|
AtMC9, metacaspase 9, AtMCP2f, metacaspase 2f, MC9, metacaspase 9, MCP2f, metacaspase 2f |
0.186 |
-0.288 |
| 57 |
256799_at |
AT3G18560
|
unknown |
0.184 |
-0.125 |
| 58 |
257670_at |
AT3G20340
|
unknown |
0.183 |
-0.010 |
| 59 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
0.183 |
0.065 |
| 60 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
0.183 |
-0.015 |
| 61 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
0.181 |
0.111 |
| 62 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.181 |
0.060 |
| 63 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.181 |
-0.051 |
| 64 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.180 |
-0.055 |
| 65 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.180 |
0.233 |
| 66 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.180 |
0.026 |
| 67 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.178 |
-0.360 |
| 68 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.177 |
0.203 |
| 69 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
0.168 |
-0.042 |
| 70 |
263379_at |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
0.166 |
-0.051 |
| 71 |
261984_at |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
0.165 |
0.084 |
| 72 |
258436_at |
AT3G16720
|
ATL2, TOXICOS EN LEVADURA 2, TL2, TOXICOS EN LEVADURA 2 |
0.163 |
0.059 |
| 73 |
259850_at |
AT1G72240
|
unknown |
0.163 |
-0.042 |
| 74 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
0.162 |
-0.022 |
| 75 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
0.161 |
0.078 |
| 76 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
0.160 |
-0.274 |
| 77 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
0.160 |
0.031 |
| 78 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
0.159 |
-0.049 |
| 79 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
0.159 |
0.056 |
| 80 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
0.159 |
0.068 |
| 81 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
0.158 |
-0.601 |
| 82 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
0.156 |
-0.111 |
| 83 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.156 |
0.046 |
| 84 |
245267_at |
AT4G14060
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.156 |
0.024 |
| 85 |
249932_at |
AT5G22390
|
unknown |
0.155 |
0.141 |
| 86 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
0.155 |
-0.088 |
| 87 |
266658_at |
AT2G25735
|
unknown |
0.155 |
-0.232 |
| 88 |
265327_at |
AT2G18210
|
unknown |
0.152 |
-0.381 |
| 89 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
0.150 |
-0.165 |
| 90 |
248961_at |
AT5G45650
|
[subtilase family protein] |
0.150 |
0.180 |
| 91 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
0.149 |
-0.014 |
| 92 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
0.149 |
-0.220 |
| 93 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.148 |
-0.125 |
| 94 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
0.147 |
0.166 |
| 95 |
247137_at |
AT5G66210
|
CPK28, calcium-dependent protein kinase 28 |
0.147 |
-0.171 |
| 96 |
265922_at |
AT2G18480
|
[Major facilitator superfamily protein] |
0.147 |
-0.034 |
| 97 |
253643_at |
AT4G29780
|
unknown |
0.147 |
0.101 |
| 98 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
0.147 |
-0.244 |
| 99 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
0.145 |
0.443 |
| 100 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
0.145 |
0.044 |
| 101 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.145 |
-0.262 |
| 102 |
260477_at |
AT1G11050
|
[Protein kinase superfamily protein] |
0.145 |
-0.075 |
| 103 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.144 |
-0.056 |
| 104 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.144 |
-0.023 |
| 105 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.144 |
-0.538 |
| 106 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
0.143 |
0.015 |
| 107 |
248726_at |
AT5G47960
|
ATRABA4C, RAB GTPase homolog A4C, RABA4C, RAB GTPase homolog A4C, SMG1, SMALL MOLECULAR WEIGHT G-PROTEIN 1 |
0.142 |
-0.451 |
| 108 |
267344_at |
AT2G44230
|
unknown |
0.142 |
-0.196 |
| 109 |
258183_at |
AT3G21550
|
AtDMP2, Arabidopsis thaliana DUF679 domain membrane protein 2, DMP2, DUF679 domain membrane protein 2 |
0.142 |
-0.097 |
| 110 |
265305_at |
AT2G20340
|
AAS, aromatic aldehyde synthase, AtAAS |
0.141 |
0.016 |
| 111 |
266396_at |
AT2G38790
|
unknown |
0.141 |
-0.163 |
| 112 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
0.140 |
0.062 |
| 113 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.139 |
-0.186 |
| 114 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
0.137 |
0.022 |
| 115 |
256185_at |
AT1G51700
|
ADOF1, DOF zinc finger protein 1, DOF1, DOF zinc finger protein 1 |
0.137 |
-0.090 |
| 116 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
0.136 |
-0.170 |
| 117 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
0.136 |
0.003 |
| 118 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.135 |
-0.138 |
| 119 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.135 |
-0.132 |
| 120 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
0.135 |
-0.190 |
| 121 |
248167_at |
AT5G54530
|
unknown |
0.135 |
0.018 |
| 122 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
0.135 |
0.224 |
| 123 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
0.134 |
-0.537 |
| 124 |
258189_at |
AT3G17860
|
JAI3, JASMONATE-INSENSITIVE 3, JAZ3, jasmonate-zim-domain protein 3, TIFY6B |
0.134 |
-0.089 |
| 125 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.133 |
0.401 |
| 126 |
262120_at |
AT1G02950
|
ATGSTF4, glutathione S-transferase F4, GST31, GLUTATHIONE S-TRANSFERASE 31, GSTF4, glutathione S-transferase F4 |
0.133 |
-0.282 |
| 127 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
0.132 |
0.095 |
| 128 |
251322_at |
AT3G61440
|
ARATH;BSAS3;1, BETA-SUBSTITUTED ALA SYNTHASE 3;1, ATCYSC1, CYSTEINE SYNTHASE C1, CYSC1, cysteine synthase C1 |
-1.030 |
0.019 |
| 129 |
253925_at |
AT4G26690
|
GDPDL3, Glycerophosphodiester phosphodiesterase (GDPD) like 3, GPDL2, GLYCEROPHOSPHODIESTERASE-LIKE 2, MRH5, MUTANT ROOT HAIR 5, SHV3, SHAVEN 3 |
-0.535 |
-0.124 |
| 130 |
250464_at |
AT5G10040
|
unknown |
-0.467 |
0.065 |
| 131 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-0.456 |
0.041 |
| 132 |
252882_at |
AT4G39675
|
unknown |
-0.343 |
-0.055 |
| 133 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.335 |
0.171 |
| 134 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
-0.330 |
-0.025 |
| 135 |
246366_at |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
-0.315 |
0.021 |
| 136 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.307 |
0.169 |
| 137 |
252511_at |
AT3G46280
|
[protein kinase-related] |
-0.294 |
-0.397 |
| 138 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.290 |
0.060 |
| 139 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
-0.285 |
0.159 |
| 140 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
-0.277 |
-0.000 |
| 141 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
-0.276 |
-0.352 |
| 142 |
252345_at |
AT3G48640
|
unknown |
-0.261 |
0.058 |
| 143 |
254574_at |
AT4G19430
|
unknown |
-0.254 |
0.092 |
| 144 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
-0.250 |
0.005 |
| 145 |
247024_at |
AT5G66985
|
unknown |
-0.237 |
0.131 |
| 146 |
254416_at |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
-0.236 |
-0.057 |
| 147 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
-0.227 |
0.015 |
| 148 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
-0.226 |
-0.011 |
| 149 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
-0.225 |
0.031 |
| 150 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
-0.225 |
-0.117 |
| 151 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.224 |
-0.168 |
| 152 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
-0.218 |
0.026 |
| 153 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.218 |
0.070 |
| 154 |
252572_at |
AT3G45290
|
ATMLO3, MILDEW RESISTANCE LOCUS O 3, MLO3, MILDEW RESISTANCE LOCUS O 3 |
-0.218 |
0.043 |
| 155 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
-0.201 |
0.212 |
| 156 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.201 |
0.124 |
| 157 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.198 |
-0.047 |
| 158 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
-0.197 |
0.054 |
| 159 |
253392_at |
AT4G32650
|
ATKC1, ARABIDOPSIS THALIANA K+ RECTIFYING CHANNEL 1, AtLKT1, A. thaliana low-K+-tolerant 1, KAT3, potassium channel in Arabidopsis thaliana 3, KC1 |
-0.197 |
-0.214 |
| 160 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
-0.196 |
-0.057 |
| 161 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
-0.196 |
0.078 |
| 162 |
260668_at |
AT1G19530
|
unknown |
-0.195 |
-0.080 |
| 163 |
249454_at |
AT5G39520
|
unknown |
-0.193 |
-0.078 |
| 164 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
-0.189 |
-0.047 |
| 165 |
249346_at |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
-0.189 |
0.011 |
| 166 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
-0.187 |
-0.131 |
| 167 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-0.185 |
-0.268 |
| 168 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
-0.185 |
0.093 |
| 169 |
256170_at |
AT1G51790
|
[Leucine-rich repeat protein kinase family protein] |
-0.183 |
-0.043 |
| 170 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-0.182 |
0.231 |
| 171 |
266797_at |
AT2G22840
|
AtGRF1, growth-regulating factor 1, GRF1, growth-regulating factor 1 |
-0.180 |
0.074 |
| 172 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
-0.180 |
0.133 |
| 173 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
-0.179 |
0.086 |
| 174 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
-0.179 |
0.097 |
| 175 |
245875_at |
AT1G26240
|
[Proline-rich extensin-like family protein] |
-0.178 |
0.091 |
| 176 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
-0.177 |
-0.022 |
| 177 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
-0.176 |
0.015 |
| 178 |
253169_at |
AT4G35120
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.176 |
0.009 |
| 179 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
-0.176 |
-0.223 |
| 180 |
248820_at |
AT5G47060
|
unknown |
-0.174 |
0.025 |
| 181 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
-0.170 |
-0.127 |
| 182 |
246415_at |
AT5G17160
|
unknown |
-0.169 |
-0.097 |
| 183 |
253506_at |
AT4G31980
|
unknown |
-0.169 |
0.162 |
| 184 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.168 |
0.157 |
| 185 |
253401_at |
AT4G32870
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.167 |
-0.233 |
| 186 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
-0.167 |
-0.027 |
| 187 |
255148_at |
AT4G08470
|
MAPKKK10, MEKK3, MAPK/ERK kinase kinase 3 |
-0.166 |
0.020 |
| 188 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.162 |
0.211 |
| 189 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
-0.162 |
0.035 |
| 190 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.161 |
0.079 |
| 191 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
-0.161 |
-0.039 |
| 192 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
-0.160 |
-0.185 |
| 193 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-0.157 |
0.066 |
| 194 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.157 |
0.373 |
| 195 |
253575_at |
AT4G31060
|
[Integrase-type DNA-binding superfamily protein] |
-0.156 |
0.301 |
| 196 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
-0.154 |
0.031 |
| 197 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
-0.152 |
0.034 |
| 198 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.150 |
-0.115 |
| 199 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-0.149 |
0.210 |
| 200 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.148 |
0.377 |
| 201 |
246388_at |
AT1G77405
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.145 |
0.060 |
| 202 |
248011_at |
AT5G56300
|
GAMT2, gibberellic acid methyltransferase 2 |
-0.144 |
0.023 |
| 203 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.144 |
0.066 |
| 204 |
248006_at |
AT5G56250
|
HAP8, HAPLESS 8 |
-0.142 |
0.082 |
| 205 |
258110_at |
AT3G14610
|
CYP72A7, cytochrome P450, family 72, subfamily A, polypeptide 7 |
-0.141 |
0.003 |
| 206 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
-0.139 |
-0.235 |
| 207 |
251633_at |
AT3G57460
|
[catalytics] |
-0.138 |
0.034 |
| 208 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
-0.137 |
-0.374 |
| 209 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
-0.137 |
0.380 |
| 210 |
259902_at |
AT1G74170
|
AtRLP13, receptor like protein 13, RLP13, receptor like protein 13 |
-0.137 |
0.155 |
| 211 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.136 |
-0.046 |
| 212 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.135 |
-0.452 |
| 213 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
-0.135 |
-0.314 |
| 214 |
248485_at |
AT5G51050
|
APC2, ATP/phosphate carrier 2 |
-0.134 |
-0.018 |
| 215 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.134 |
0.212 |
| 216 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.133 |
0.043 |
| 217 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.133 |
0.105 |
| 218 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
-0.133 |
-0.151 |
| 219 |
264635_at |
AT1G65500
|
unknown |
-0.132 |
-0.190 |
| 220 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
-0.132 |
0.074 |
| 221 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.131 |
-0.173 |
| 222 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
-0.130 |
0.128 |
| 223 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
-0.130 |
-0.238 |
| 224 |
254253_at |
AT4G23320
|
CRK24, cysteine-rich RLK (RECEPTOR-like protein kinase) 24 |
-0.129 |
0.224 |
| 225 |
255045_at |
AT4G09690
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.128 |
-0.026 |
| 226 |
245861_at |
AT5G28300
|
AtGT2L, GT2L, GT-2Like protein |
-0.128 |
0.368 |
| 227 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
-0.127 |
-0.015 |
| 228 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
-0.126 |
-0.285 |
| 229 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.126 |
-0.139 |
| 230 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-0.126 |
-0.192 |
| 231 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.125 |
-0.094 |
| 232 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
-0.123 |
-0.002 |
| 233 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
-0.123 |
-0.284 |
| 234 |
253887_at |
AT4G27730
|
ATOPT6, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 6, OPT6, oligopeptide transporter 1 |
-0.123 |
0.083 |
| 235 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
-0.123 |
-0.137 |
| 236 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
-0.122 |
0.071 |
| 237 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
-0.122 |
-0.101 |
| 238 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
-0.122 |
0.006 |
| 239 |
245968_at |
AT5G19810
|
[Proline-rich extensin-like family protein] |
-0.122 |
0.015 |
| 240 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
-0.122 |
0.001 |
| 241 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.122 |
-0.111 |
| 242 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
-0.121 |
-0.305 |
| 243 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
-0.121 |
0.004 |
| 244 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
-0.121 |
-0.195 |
| 245 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.121 |
0.116 |
| 246 |
248089_at |
AT5G55080
|
AtRAN4, ras-related nuclear protein 4, RAN4, ras-related nuclear protein 4 |
-0.121 |
0.020 |
| 247 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
-0.120 |
0.182 |
| 248 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
-0.119 |
-0.121 |
| 249 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
-0.119 |
-0.035 |
| 250 |
249871_at |
AT5G23110
|
[Zinc finger, C3HC4 type (RING finger) family protein] |
-0.118 |
0.054 |
| 251 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
-0.117 |
-0.038 |
| 252 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
-0.117 |
-0.026 |
| 253 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.116 |
-0.004 |