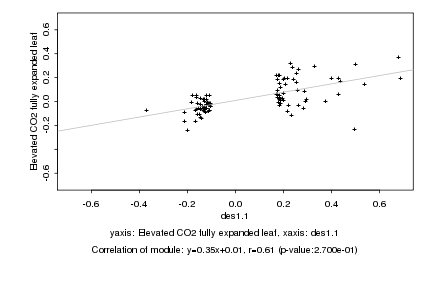
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
des1.1 |
Log2 signal ratio
Elevated CO2 fully expanded leaf |
| 1 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
0.686 |
0.198 |
| 2 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.679 |
0.374 |
| 3 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.534 |
0.149 |
| 4 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.496 |
0.311 |
| 5 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.494 |
-0.227 |
| 6 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.435 |
0.169 |
| 7 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.428 |
0.195 |
| 8 |
256569_at |
AT3G19550
|
unknown |
0.427 |
0.061 |
| 9 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
0.400 |
0.197 |
| 10 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.374 |
0.004 |
| 11 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.329 |
0.298 |
| 12 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.293 |
0.017 |
| 13 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.289 |
0.003 |
| 14 |
249454_at |
AT5G39520
|
unknown |
0.285 |
0.091 |
| 15 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.283 |
-0.051 |
| 16 |
247610_at |
AT5G60630
|
unknown |
0.261 |
-0.029 |
| 17 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.259 |
0.275 |
| 18 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.258 |
0.094 |
| 19 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
0.253 |
0.240 |
| 20 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.252 |
0.166 |
| 21 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.240 |
0.192 |
| 22 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.234 |
0.291 |
| 23 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.231 |
-0.113 |
| 24 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.226 |
0.320 |
| 25 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
0.219 |
-0.025 |
| 26 |
263829_at |
AT2G40435
|
unknown |
0.215 |
-0.077 |
| 27 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.213 |
0.197 |
| 28 |
250723_at |
AT5G06300
|
LOG7, LONELY GUY 7 |
0.206 |
0.147 |
| 29 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.202 |
0.200 |
| 30 |
251304_at |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
0.198 |
0.075 |
| 31 |
258791_at |
AT3G04720
|
AtPR4, HEL, HEVEIN-LIKE, PR-4, PR4, pathogenesis-related 4 |
0.197 |
0.189 |
| 32 |
259103_at |
AT3G11690
|
unknown |
0.197 |
0.015 |
| 33 |
263046_at |
AT2G05380
|
GRP3S, glycine-rich protein 3 short isoform |
0.193 |
0.028 |
| 34 |
253224_at |
AT4G34860
|
A/N-InvB, alkaline/neutral invertase B |
0.185 |
0.009 |
| 35 |
247280_at |
AT5G64260
|
EXL2, EXORDIUM like 2 |
0.185 |
0.122 |
| 36 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.184 |
0.015 |
| 37 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.184 |
-0.012 |
| 38 |
249839_at |
AT5G23405
|
[HMG-box (high mobility group) DNA-binding family protein] |
0.183 |
0.038 |
| 39 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.183 |
0.017 |
| 40 |
265341_at |
AT2G18360
|
[alpha/beta-Hydrolases superfamily protein] |
0.182 |
0.029 |
| 41 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.182 |
0.222 |
| 42 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.181 |
-0.026 |
| 43 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
0.181 |
0.054 |
| 44 |
252993_at |
AT4G38540
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.180 |
0.155 |
| 45 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.179 |
0.218 |
| 46 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
0.177 |
-0.005 |
| 47 |
260897_at |
AT1G29330
|
AERD2, ARABIDOPSIS ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ATERD2, ARABIDOPSIS THALIANA ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ERD2, ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2 |
0.173 |
0.066 |
| 48 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.173 |
0.192 |
| 49 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.172 |
0.097 |
| 50 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
0.172 |
0.035 |
| 51 |
265382_at |
AT2G16790
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.171 |
0.062 |
| 52 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.171 |
0.225 |
| 53 |
258017_at |
AT3G19370
|
unknown |
-0.373 |
-0.074 |
| 54 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.216 |
-0.087 |
| 55 |
247469_at |
AT5G62165
|
AGL42, AGAMOUS-like 42, FYF, FOREVER YOUNG FLOWER |
-0.215 |
-0.167 |
| 56 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.200 |
-0.240 |
| 57 |
266971_at |
AT2G39580
|
unknown |
-0.187 |
-0.004 |
| 58 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.179 |
0.052 |
| 59 |
244997_at |
ATCG00170
|
RPOC2 |
-0.169 |
-0.071 |
| 60 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
-0.169 |
-0.163 |
| 61 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
-0.166 |
0.055 |
| 62 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.165 |
0.035 |
| 63 |
262773_at |
AT1G13220
|
CRWN2, CROWDED NUCLEI 2, LINC2, LITTLE NUCLEI2 |
-0.164 |
-0.059 |
| 64 |
244995_at |
ATCG00150
|
ATPI |
-0.161 |
-0.107 |
| 65 |
245522_at |
AT4G15890
|
[binding] |
-0.159 |
-0.012 |
| 66 |
263384_at |
AT2G40130
|
SMXL8, SMAX1-like 8 |
-0.154 |
-0.058 |
| 67 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
-0.151 |
-0.105 |
| 68 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.149 |
-0.131 |
| 69 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
-0.148 |
-0.053 |
| 70 |
259282_at |
AT3G11430
|
ATGPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5, GPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5 |
-0.148 |
-0.017 |
| 71 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.148 |
0.030 |
| 72 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.147 |
-0.063 |
| 73 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.143 |
-0.140 |
| 74 |
258888_at |
AT3G05620
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.140 |
-0.036 |
| 75 |
258661_at |
AT3G02930
|
unknown |
-0.136 |
-0.053 |
| 76 |
258918_at |
AT3G10560
|
CYP77A7, CYTOCHROME P450, FAMILY 77, SUBFAMILY A, POLYPEPTIDE 7, UNE9, UNFERTILIZED EMBRYO SAC 9 |
-0.136 |
0.019 |
| 77 |
255876_at |
AT2G40480
|
unknown |
-0.134 |
-0.068 |
| 78 |
254200_at |
AT4G24110
|
unknown |
-0.133 |
-0.052 |
| 79 |
253947_at |
AT4G26760
|
MAP65-2, microtubule-associated protein 65-2 |
-0.131 |
-0.048 |
| 80 |
260937_at |
AT1G45160
|
[Protein kinase superfamily protein] |
-0.131 |
0.004 |
| 81 |
254710_at |
AT4G18050
|
ABCB9, ATP-binding cassette B9, PGP9, P-glycoprotein 9 |
-0.130 |
-0.058 |
| 82 |
247397_at |
AT5G62940
|
DOF5.6, DNA BINDING WITH ONE FINGER 5.6, HCA2, HIGH CAMBIAL ACTIVITY2 |
-0.130 |
-0.076 |
| 83 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.129 |
0.017 |
| 84 |
248441_at |
AT5G51270
|
[U-box domain-containing protein kinase family protein] |
-0.128 |
-0.052 |
| 85 |
253082_at |
AT4G36230
|
unknown |
-0.125 |
-0.089 |
| 86 |
255664_at |
AT4G00440
|
TRM15, TON1 Recruiting Motif 15 |
-0.124 |
-0.053 |
| 87 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.124 |
0.051 |
| 88 |
253849_at |
AT4G28080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.123 |
-0.057 |
| 89 |
262921_at |
AT1G79430
|
APL, ALTERED PHLOEM DEVELOPMENT, WDY, WOODY |
-0.123 |
-0.021 |
| 90 |
262391_at |
AT1G49530
|
GGPS6, geranylgeranyl pyrophosphate synthase 6 |
-0.122 |
-0.024 |
| 91 |
262577_at |
AT1G15290
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.121 |
-0.052 |
| 92 |
263882_at |
AT2G21790
|
ATRNR1, RIBONUCLEOTIDE REDUCTASE LARGE SUBUNIT 1, CLS8, CRINKLY LEAVES 8, DPD2, defective in pollen organelle DNA degradation 2, R1, RIBONUCLEOTIDE REDUCTASE 1, RNR1, ribonucleotide reductase 1 |
-0.121 |
0.018 |
| 93 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
-0.117 |
-0.002 |
| 94 |
263492_at |
AT2G42560
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
-0.116 |
-0.009 |
| 95 |
266990_at |
AT2G39190
|
ATATH8 |
-0.113 |
-0.080 |
| 96 |
256603_at |
AT3G28270
|
unknown |
-0.112 |
-0.072 |
| 97 |
261427_at |
AT1G18670
|
IBS1, IMPAIRED IN BABA-INDUCED STERILITY 1 |
-0.110 |
-0.009 |
| 98 |
258898_at |
AT3G05740
|
RECQI1, RECQ helicase l1 |
-0.109 |
0.052 |
| 99 |
267021_at |
AT2G39300
|
unknown |
-0.109 |
-0.071 |
| 100 |
267410_at |
AT2G34920
|
EDA18, embryo sac development arrest 18 |
-0.109 |
-0.032 |
| 101 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.105 |
-0.035 |
| 102 |
258123_at |
AT3G18040
|
MPK9, MAP kinase 9 |
-0.105 |
-0.018 |
| 103 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
-0.105 |
-0.013 |
| 104 |
255053_at |
AT4G09730
|
RH39, RH39 |
-0.104 |
-0.038 |