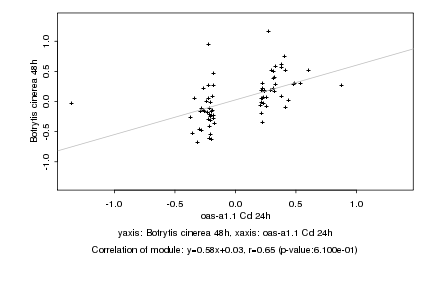
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
oas-a1.1 Cd 24h |
Log2 signal ratio
Botrytis cinerea 48h |
| 1 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
0.870 |
0.269 |
| 2 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.596 |
0.523 |
| 3 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.531 |
0.304 |
| 4 |
253832_at |
AT4G27654
|
unknown |
0.485 |
0.310 |
| 5 |
253799_at |
AT4G28140
|
[Integrase-type DNA-binding superfamily protein] |
0.474 |
0.288 |
| 6 |
260688_at |
AT1G17665
|
unknown |
0.433 |
0.032 |
| 7 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.413 |
0.520 |
| 8 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.412 |
-0.084 |
| 9 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.403 |
0.750 |
| 10 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.375 |
0.623 |
| 11 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.375 |
0.094 |
| 12 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.374 |
0.566 |
| 13 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.325 |
0.291 |
| 14 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.323 |
0.590 |
| 15 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
0.318 |
0.406 |
| 16 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.317 |
0.180 |
| 17 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.311 |
0.385 |
| 18 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.310 |
0.227 |
| 19 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.308 |
0.500 |
| 20 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.290 |
0.520 |
| 21 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
0.288 |
0.188 |
| 22 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
0.272 |
1.169 |
| 23 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.251 |
0.071 |
| 24 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
0.249 |
-0.068 |
| 25 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.238 |
0.168 |
| 26 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
0.236 |
0.199 |
| 27 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
0.230 |
-0.025 |
| 28 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.224 |
0.072 |
| 29 |
260237_at |
AT1G74430
|
ATMYB95, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 95, ATMYBCP66, ARABIDOPSIS THALIANA MYB DOMAIN CONTAINING PROTEIN 66, MYB95, myb domain protein 95 |
0.223 |
0.052 |
| 30 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.222 |
-0.335 |
| 31 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.221 |
0.302 |
| 32 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
0.220 |
0.227 |
| 33 |
253322_at |
AT4G33980
|
unknown |
0.211 |
0.060 |
| 34 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.209 |
-0.191 |
| 35 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
0.208 |
-0.006 |
| 36 |
251640_at |
AT3G57450
|
unknown |
0.208 |
0.199 |
| 37 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.205 |
-0.061 |
| 38 |
245286_at |
AT4G14880
|
ATCYS-3A, CYTACS1, OASA1, O-acetylserine (thiol) lyase (OAS-TL) isoform A1, OLD3, ONSET OF LEAF DEATH 3 |
-1.361 |
-0.024 |
| 39 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.378 |
-0.258 |
| 40 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.355 |
-0.525 |
| 41 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
-0.342 |
0.061 |
| 42 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.319 |
-0.677 |
| 43 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.304 |
-0.455 |
| 44 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.296 |
-0.162 |
| 45 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.288 |
-0.469 |
| 46 |
247324_at |
AT5G64190
|
unknown |
-0.287 |
-0.113 |
| 47 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.268 |
-0.146 |
| 48 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
-0.267 |
0.218 |
| 49 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.257 |
-0.157 |
| 50 |
266824_at |
AT2G22800
|
HAT9 |
-0.246 |
0.015 |
| 51 |
266363_at |
AT2G41250
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.236 |
-0.180 |
| 52 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.228 |
0.059 |
| 53 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.227 |
0.272 |
| 54 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
-0.226 |
-0.290 |
| 55 |
251743_at |
AT3G55890
|
[Yippee family putative zinc-binding protein] |
-0.223 |
0.948 |
| 56 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.218 |
-0.598 |
| 57 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.216 |
-0.203 |
| 58 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
-0.216 |
-0.108 |
| 59 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.216 |
-0.403 |
| 60 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.215 |
-0.610 |
| 61 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.213 |
-0.243 |
| 62 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.209 |
-0.546 |
| 63 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
-0.207 |
-0.009 |
| 64 |
255824_at |
AT2G40530
|
unknown |
-0.207 |
-0.299 |
| 65 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
-0.206 |
-0.618 |
| 66 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.204 |
-0.226 |
| 67 |
252501_at |
AT3G46880
|
unknown |
-0.201 |
-0.150 |
| 68 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.194 |
0.086 |
| 69 |
261567_at |
AT1G33055
|
unknown |
-0.191 |
-0.134 |
| 70 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
-0.186 |
-0.272 |
| 71 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
-0.186 |
0.273 |
| 72 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
-0.186 |
0.468 |
| 73 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.184 |
-0.217 |
| 74 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.177 |
-0.352 |