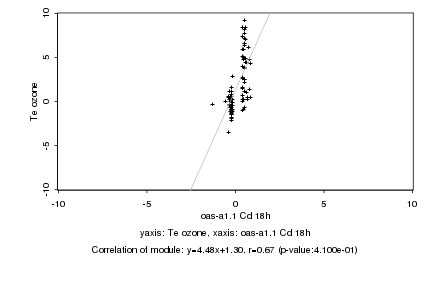
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
oas-a1.1 Cd 18h |
Log2 signal ratio
Te ozone |
| 1 |
250702_at |
AT5G06730
|
[Peroxidase superfamily protein] |
0.833 |
0.544 |
| 2 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.809 |
4.360 |
| 3 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.792 |
4.802 |
| 4 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.783 |
1.435 |
| 5 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.697 |
6.151 |
| 6 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.660 |
0.329 |
| 7 |
245226_at |
AT3G29970
|
[B12D protein] |
0.629 |
0.471 |
| 8 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.579 |
1.029 |
| 9 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.570 |
4.504 |
| 10 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.558 |
8.420 |
| 11 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.540 |
4.940 |
| 12 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.518 |
7.043 |
| 13 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
0.514 |
4.468 |
| 14 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.506 |
6.407 |
| 15 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.502 |
3.946 |
| 16 |
251824_at |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
0.499 |
-0.601 |
| 17 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.494 |
4.975 |
| 18 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.491 |
3.798 |
| 19 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.488 |
7.758 |
| 20 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.486 |
1.214 |
| 21 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.484 |
9.301 |
| 22 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.481 |
6.648 |
| 23 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.481 |
8.270 |
| 24 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.480 |
2.582 |
| 25 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.478 |
7.194 |
| 26 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.475 |
2.585 |
| 27 |
253067_at |
AT4G37780
|
ATMYB87, MYB87, myb domain protein 87 |
0.462 |
2.237 |
| 28 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.460 |
4.963 |
| 29 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.449 |
5.990 |
| 30 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.415 |
0.268 |
| 31 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
0.415 |
4.836 |
| 32 |
252014_at |
AT3G52870
|
[IQ calmodulin-binding motif family protein] |
0.412 |
0.236 |
| 33 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
0.409 |
-0.882 |
| 34 |
264774_at |
AT1G22890
|
unknown |
0.399 |
5.059 |
| 35 |
249941_at |
AT5G22270
|
unknown |
0.389 |
2.681 |
| 36 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.388 |
8.495 |
| 37 |
252862_at |
AT4G39830
|
[Cupredoxin superfamily protein] |
0.384 |
5.202 |
| 38 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.371 |
1.604 |
| 39 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.369 |
7.480 |
| 40 |
247922_at |
AT5G57500
|
[Galactosyltransferase family protein] |
0.369 |
2.752 |
| 41 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.369 |
3.997 |
| 42 |
264842_at |
AT1G03700
|
[Uncharacterised protein family (UPF0497)] |
0.359 |
0.238 |
| 43 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.356 |
0.778 |
| 44 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.352 |
6.000 |
| 45 |
249767_at |
AT5G24090
|
ATCHIA, chitinase A, CHIA, chitinase A |
0.352 |
-0.991 |
| 46 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.346 |
0.045 |
| 47 |
255999_at |
AT1G29860
|
ATWRKY71, WRKY DNA-BINDING PROTEIN 71, WRKY71, WRKY DNA-binding protein 71 |
0.345 |
1.502 |
| 48 |
245286_at |
AT4G14880
|
ATCYS-3A, CYTACS1, OASA1, O-acetylserine (thiol) lyase (OAS-TL) isoform A1, OLD3, ONSET OF LEAF DEATH 3 |
-1.306 |
-0.288 |
| 49 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
-0.605 |
0.013 |
| 50 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
-0.420 |
0.626 |
| 51 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.410 |
-3.452 |
| 52 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.402 |
0.544 |
| 53 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.387 |
-0.451 |
| 54 |
261466_at |
AT1G07690
|
unknown |
-0.372 |
1.163 |
| 55 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.350 |
-0.328 |
| 56 |
249203_at |
AT5G42590
|
CYP71A16, cytochrome P450, family 71, subfamily A, polypeptide 16, MRO, marneral oxidase |
-0.345 |
0.238 |
| 57 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.332 |
0.231 |
| 58 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.327 |
0.016 |
| 59 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.310 |
-0.618 |
| 60 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.304 |
-1.100 |
| 61 |
252536_at |
AT3G45700
|
[Major facilitator superfamily protein] |
-0.304 |
0.616 |
| 62 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.299 |
-0.650 |
| 63 |
258912_at |
AT3G06460
|
[GNS1/SUR4 membrane protein family] |
-0.293 |
0.238 |
| 64 |
251829_at |
AT3G55020
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
-0.271 |
-0.356 |
| 65 |
253525_at |
AT4G31330
|
unknown |
-0.270 |
-1.435 |
| 66 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.269 |
-0.673 |
| 67 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
-0.260 |
-1.763 |
| 68 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
-0.259 |
-0.889 |
| 69 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
-0.259 |
-0.415 |
| 70 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.257 |
-1.387 |
| 71 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.256 |
-1.103 |
| 72 |
251289_at |
AT3G61830
|
ARF18, auxin response factor 18 |
-0.251 |
-1.132 |
| 73 |
256600_at |
AT3G14850
|
TBL41, TRICHOME BIREFRINGENCE-LIKE 41 |
-0.251 |
0.847 |
| 74 |
258676_at |
AT3G08600
|
unknown |
-0.251 |
-2.090 |
| 75 |
263226_at |
AT1G30690
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.251 |
-0.842 |
| 76 |
254510_at |
AT4G20210
|
TPS08, terpene synthase 8 |
-0.249 |
0.238 |
| 77 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-0.248 |
-1.900 |
| 78 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.248 |
1.163 |
| 79 |
260969_at |
AT1G12240
|
ATBETAFRUCT4, AtFRUCT4, AtVI2, FRUCT4, fructosidase 4, VAC-INV, VACUOLAR INVERTASE, VI2, vacuolar invertase 2 |
-0.248 |
-1.336 |
| 80 |
260558_at |
AT2G43600
|
[Chitinase family protein] |
-0.236 |
0.238 |
| 81 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.235 |
-0.848 |
| 82 |
254467_at |
AT4G20450
|
[Leucine-rich repeat protein kinase family protein] |
-0.230 |
0.238 |
| 83 |
254344_at |
AT4G22110
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.228 |
0.616 |
| 84 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.226 |
-0.919 |
| 85 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.225 |
1.591 |
| 86 |
265049_at |
AT1G52060
|
[Mannose-binding lectin superfamily protein] |
-0.223 |
0.238 |
| 87 |
258332_at |
AT3G16180
|
NRT1.12, nitrate transporter 1.12 |
-0.223 |
0.908 |
| 88 |
265103_at |
AT1G31070
|
GlcNAc1pUT1, N-acetylglucosamine-1-phosphate uridylyltransferase 1 |
-0.221 |
-0.866 |
| 89 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
-0.220 |
2.854 |
| 90 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.220 |
-1.069 |
| 91 |
248652_at |
AT5G49270
|
COBL9, COBRA-LIKE 9, DER9, DEFORMED ROOT HAIRS 9, MRH4, MUTANT ROOT HAIR 4, SHV2, SHAVEN 2 |
-0.217 |
0.238 |
| 92 |
250717_at |
AT5G06200
|
CASP4, Casparian strip membrane domain protein 4 |
-0.217 |
-0.388 |
| 93 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
-0.216 |
-0.051 |