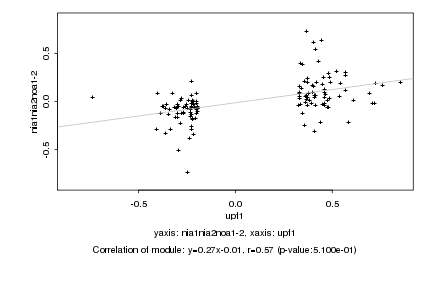
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
upf1 |
Log2 signal ratio
nia1nia2noa1-2 |
| 1 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
0.851 |
0.201 |
| 2 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
0.755 |
0.170 |
| 3 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.721 |
0.188 |
| 4 |
254252_at |
AT4G23310
|
CRK23, cysteine-rich RLK (RECEPTOR-like protein kinase) 23 |
0.714 |
-0.016 |
| 5 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
0.706 |
-0.016 |
| 6 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
0.692 |
0.091 |
| 7 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
0.608 |
0.012 |
| 8 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
0.584 |
-0.216 |
| 9 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.567 |
0.275 |
| 10 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
0.566 |
0.119 |
| 11 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.565 |
0.306 |
| 12 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.541 |
0.189 |
| 13 |
252345_at |
AT3G48640
|
unknown |
0.537 |
0.061 |
| 14 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
0.519 |
0.314 |
| 15 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.490 |
0.199 |
| 16 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
0.484 |
0.039 |
| 17 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.484 |
0.258 |
| 18 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.480 |
0.292 |
| 19 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.478 |
-0.055 |
| 20 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.475 |
-0.054 |
| 21 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.472 |
0.015 |
| 22 |
252383_at |
AT3G47780
|
ABCA7, ATP-binding cassette A7, ATATH6, A. THALIANA ABC2 HOMOLOG 6, ATH6, ABC2 homolog 6 |
0.462 |
0.082 |
| 23 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
0.460 |
0.135 |
| 24 |
251419_at |
AT3G60470
|
unknown |
0.459 |
-0.038 |
| 25 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.457 |
0.046 |
| 26 |
263854_at |
AT2G04430
|
atnudt5, nudix hydrolase homolog 5, NUDT5, nudix hydrolase homolog 5 |
0.456 |
-0.018 |
| 27 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.456 |
0.101 |
| 28 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.450 |
0.252 |
| 29 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
0.449 |
0.182 |
| 30 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.447 |
-0.023 |
| 31 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.440 |
0.636 |
| 32 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
0.436 |
-0.209 |
| 33 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.425 |
0.416 |
| 34 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.416 |
0.201 |
| 35 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.412 |
-0.038 |
| 36 |
257591_at |
AT3G24900
|
AtRLP39, receptor like protein 39, RLP39, receptor like protein 39 |
0.412 |
0.068 |
| 37 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.410 |
0.546 |
| 38 |
249896_at |
AT5G22530
|
unknown |
0.408 |
0.065 |
| 39 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.407 |
0.049 |
| 40 |
265892_at |
AT2G15020
|
unknown |
0.406 |
-0.309 |
| 41 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.400 |
0.620 |
| 42 |
255259_at |
AT4G05020
|
NDB2, NAD(P)H dehydrogenase B2 |
0.400 |
0.164 |
| 43 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
0.398 |
0.103 |
| 44 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.394 |
0.168 |
| 45 |
254229_at |
AT4G23610
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.389 |
-0.014 |
| 46 |
259009_at |
AT3G09260
|
BGLU23, LEB, LONG ER BODY, PSR3.1, PYK10 |
0.382 |
0.018 |
| 47 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.374 |
0.088 |
| 48 |
259850_at |
AT1G72240
|
unknown |
0.371 |
-0.033 |
| 49 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.370 |
0.245 |
| 50 |
251422_at |
AT3G60540
|
[Preprotein translocase Sec, Sec61-beta subunit protein] |
0.369 |
0.043 |
| 51 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
0.368 |
0.204 |
| 52 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.366 |
0.737 |
| 53 |
251059_at |
AT5G01810
|
ATPK10, PROTEIN KINASE 10, CIPK15, CBL-interacting protein kinase 15, PKS3, SIP2, SOS3-INTERACTING PROTEIN 2, SNRK3.1, SNF1-RELATED PROTEIN KINASE 3.1 |
0.365 |
0.030 |
| 54 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.363 |
0.067 |
| 55 |
254253_at |
AT4G23320
|
CRK24, cysteine-rich RLK (RECEPTOR-like protein kinase) 24 |
0.359 |
-0.005 |
| 56 |
254390_at |
AT4G21940
|
CPK15, calcium-dependent protein kinase 15 |
0.358 |
0.057 |
| 57 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.355 |
-0.242 |
| 58 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.355 |
0.214 |
| 59 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.343 |
-0.124 |
| 60 |
246125_at |
AT5G19875
|
unknown |
0.342 |
0.389 |
| 61 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.339 |
0.137 |
| 62 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.336 |
-0.028 |
| 63 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.332 |
0.397 |
| 64 |
245375_at |
AT4G17660
|
[Protein kinase superfamily protein] |
0.331 |
0.055 |
| 65 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
0.328 |
0.037 |
| 66 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.328 |
0.160 |
| 67 |
256188_at |
AT1G30160
|
unknown |
0.326 |
0.102 |
| 68 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.326 |
0.093 |
| 69 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.325 |
-0.041 |
| 70 |
248816_at |
AT5G47010
|
ATUPF1, LBA1, LOW-LEVEL BETA-AMYLASE 1, UPF1 |
-0.742 |
0.048 |
| 71 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
-0.412 |
-0.283 |
| 72 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
-0.403 |
0.090 |
| 73 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.388 |
-0.124 |
| 74 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
-0.378 |
-0.046 |
| 75 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.376 |
-0.046 |
| 76 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.364 |
-0.063 |
| 77 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
-0.363 |
-0.327 |
| 78 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.359 |
-0.021 |
| 79 |
261175_at |
AT1G04800
|
[glycine-rich protein] |
-0.347 |
-0.127 |
| 80 |
261203_at |
AT1G12845
|
unknown |
-0.343 |
-0.075 |
| 81 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-0.337 |
-0.290 |
| 82 |
262040_at |
AT1G80080
|
AtRLP17, Receptor Like Protein 17, TMM, TOO MANY MOUTHS |
-0.328 |
0.084 |
| 83 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.317 |
-0.060 |
| 84 |
248509_at |
AT5G50335
|
unknown |
-0.310 |
-0.160 |
| 85 |
248420_at |
AT5G51560
|
[Leucine-rich repeat protein kinase family protein] |
-0.306 |
-0.069 |
| 86 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.304 |
-0.156 |
| 87 |
245789_at |
AT1G32090
|
[early-responsive to dehydration stress protein (ERD4)] |
-0.303 |
-0.046 |
| 88 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.300 |
-0.025 |
| 89 |
247048_at |
AT5G66560
|
[Phototropic-responsive NPH3 family protein] |
-0.300 |
-0.121 |
| 90 |
249807_at |
AT5G23870
|
[Pectinacetylesterase family protein] |
-0.297 |
-0.048 |
| 91 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.296 |
-0.498 |
| 92 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-0.287 |
0.013 |
| 93 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.286 |
-0.221 |
| 94 |
267144_at |
AT2G38110
|
ATGPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6, GPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6 |
-0.282 |
-0.121 |
| 95 |
250936_at |
AT5G03120
|
unknown |
-0.279 |
0.041 |
| 96 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.271 |
-0.114 |
| 97 |
258796_at |
AT3G04630
|
WDL1, WVD2-like 1 |
-0.270 |
-0.057 |
| 98 |
260081_at |
AT1G78170
|
unknown |
-0.269 |
-0.109 |
| 99 |
258376_at |
AT3G17680
|
[Kinase interacting (KIP1-like) family protein] |
-0.261 |
-0.049 |
| 100 |
258530_at |
AT3G06840
|
unknown |
-0.257 |
-0.022 |
| 101 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.251 |
-0.070 |
| 102 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.248 |
-0.727 |
| 103 |
266035_at |
AT2G05990
|
ENR1, ENOYL-ACP REDUCTASE 1, MOD1, MOSAIC DEATH 1 |
-0.244 |
0.018 |
| 104 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
-0.239 |
-0.375 |
| 105 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-0.237 |
-0.107 |
| 106 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.236 |
-0.150 |
| 107 |
252280_at |
AT3G49260
|
iqd21, IQ-domain 21 |
-0.233 |
-0.075 |
| 108 |
255675_at |
AT4G00480
|
ATMYC1, myc1 |
-0.232 |
-0.016 |
| 109 |
259180_at |
AT3G01680
|
AtSEOR1, Arabidopsis thaliana sieve element occlusion-related 1, SEOb, sieve element occlusion b, SEOR1, Sieve-Element-Occlusion-Related 1 |
-0.231 |
-0.128 |
| 110 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
-0.231 |
-0.047 |
| 111 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
-0.230 |
0.215 |
| 112 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.230 |
-0.058 |
| 113 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
-0.230 |
-0.288 |
| 114 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
-0.230 |
0.066 |
| 115 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.229 |
-0.257 |
| 116 |
264529_at |
AT1G30820
|
[CTP synthase family protein] |
-0.226 |
-0.168 |
| 117 |
246097_at |
AT5G20270
|
HHP1, heptahelical transmembrane protein1 |
-0.225 |
0.005 |
| 118 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.225 |
-0.178 |
| 119 |
255709_at |
AT4G00180
|
YAB3, YABBY3 |
-0.224 |
0.013 |
| 120 |
248427_at |
AT5G51750
|
ATSBT1.3, subtilase 1.3, SBT1.3, subtilase 1.3 |
-0.223 |
-0.011 |
| 121 |
263628_at |
AT2G04780
|
FLA7, FASCICLIN-like arabinoogalactan 7 |
-0.221 |
-0.026 |
| 122 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.219 |
0.004 |
| 123 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.217 |
-0.335 |
| 124 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
-0.214 |
-0.057 |
| 125 |
258663_at |
AT3G08670
|
unknown |
-0.212 |
-0.061 |
| 126 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
-0.211 |
-0.063 |
| 127 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.208 |
-0.168 |
| 128 |
266946_at |
AT2G18890
|
[Protein kinase superfamily protein] |
-0.206 |
-0.114 |
| 129 |
258222_at |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.205 |
-0.020 |
| 130 |
251881_at |
AT3G54250
|
[GHMP kinase family protein] |
-0.204 |
-0.078 |
| 131 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
-0.204 |
0.006 |
| 132 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.202 |
0.085 |
| 133 |
259151_at |
AT3G10310
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.200 |
-0.065 |
| 134 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-0.200 |
-0.051 |
| 135 |
246288_at |
AT1G31850
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.200 |
-0.067 |
| 136 |
261169_at |
AT1G04920
|
ATSPS3F, sucrose phosphate synthase 3F, SPS3F, sucrose phosphate synthase 3F |
-0.199 |
-0.054 |
| 137 |
255851_at |
AT1G67040
|
TRM22, TON1 Recruiting Motif 22 |
-0.199 |
-0.094 |