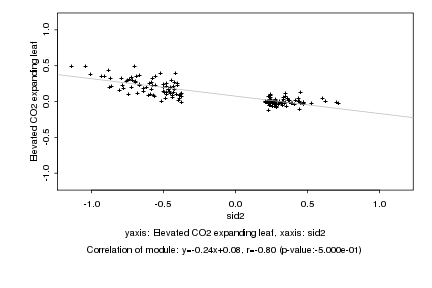
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
sid2 |
Log2 signal ratio
Elevated CO2 expanding leaf |
| 1 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
0.714 |
-0.016 |
| 2 |
246200_at |
AT4G37240
|
unknown |
0.699 |
-0.007 |
| 3 |
264238_at |
AT1G54740
|
unknown |
0.625 |
0.003 |
| 4 |
257076_at |
AT3G19680
|
unknown |
0.598 |
0.047 |
| 5 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
0.526 |
-0.014 |
| 6 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
0.471 |
-0.007 |
| 7 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.469 |
-0.006 |
| 8 |
263829_at |
AT2G40435
|
unknown |
0.468 |
-0.040 |
| 9 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.468 |
-0.025 |
| 10 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.450 |
0.137 |
| 11 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.447 |
-0.026 |
| 12 |
255943_at |
AT1G22370
|
AtUGT85A5, UDP-glucosyl transferase 85A5, UGT85A5, UDP-glucosyl transferase 85A5 |
0.443 |
-0.111 |
| 13 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.441 |
0.004 |
| 14 |
252014_at |
AT3G52870
|
[IQ calmodulin-binding motif family protein] |
0.436 |
0.011 |
| 15 |
248432_at |
AT5G51390
|
unknown |
0.434 |
0.046 |
| 16 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
0.415 |
0.027 |
| 17 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.406 |
-0.038 |
| 18 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
0.392 |
-0.025 |
| 19 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
0.383 |
-0.016 |
| 20 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
0.375 |
-0.011 |
| 21 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
0.371 |
-0.011 |
| 22 |
260367_at |
AT1G69760
|
unknown |
0.369 |
0.028 |
| 23 |
246253_at |
AT4G37260
|
ATMYB73, MYB73, myb domain protein 73 |
0.363 |
0.015 |
| 24 |
264379_at |
AT2G25200
|
unknown |
0.359 |
0.053 |
| 25 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.349 |
-0.063 |
| 26 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.345 |
-0.008 |
| 27 |
266259_at |
AT2G27830
|
unknown |
0.344 |
0.123 |
| 28 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.344 |
0.081 |
| 29 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
0.336 |
-0.024 |
| 30 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
0.331 |
0.031 |
| 31 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.329 |
0.067 |
| 32 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.326 |
0.006 |
| 33 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
0.324 |
-0.055 |
| 34 |
262245_at |
AT1G48240
|
ATNPSN12, NPSN12, novel plant snare 12 |
0.319 |
-0.026 |
| 35 |
253165_at |
AT4G35320
|
unknown |
0.317 |
-0.016 |
| 36 |
247585_at |
AT5G60680
|
unknown |
0.305 |
0.012 |
| 37 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
0.302 |
-0.009 |
| 38 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
0.295 |
-0.035 |
| 39 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.284 |
0.001 |
| 40 |
257895_at |
AT3G16950
|
LPD1, lipoamide dehydrogenase 1, ptlpd1 |
0.284 |
-0.076 |
| 41 |
261921_at |
AT1G65900
|
unknown |
0.281 |
-0.063 |
| 42 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
0.277 |
0.033 |
| 43 |
248776_at |
AT5G47900
|
unknown |
0.274 |
-0.041 |
| 44 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
0.274 |
-0.020 |
| 45 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
0.273 |
-0.027 |
| 46 |
259996_at |
AT1G67910
|
unknown |
0.272 |
0.010 |
| 47 |
250233_at |
AT5G13460
|
IQD11, IQ-domain 11 |
0.272 |
0.016 |
| 48 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
0.272 |
-0.036 |
| 49 |
264987_at |
AT1G27030
|
unknown |
0.270 |
-0.064 |
| 50 |
252230_at |
AT3G49810
|
[ARM repeat superfamily protein] |
0.268 |
-0.019 |
| 51 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.267 |
-0.041 |
| 52 |
250937_at |
AT5G03230
|
unknown |
0.264 |
-0.011 |
| 53 |
263421_at |
AT2G17230
|
EXL5, EXORDIUM like 5 |
0.264 |
-0.008 |
| 54 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.262 |
0.001 |
| 55 |
257672_at |
AT3G20300
|
unknown |
0.256 |
-0.022 |
| 56 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
0.256 |
0.007 |
| 57 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
0.251 |
-0.064 |
| 58 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
0.249 |
0.025 |
| 59 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
0.241 |
0.099 |
| 60 |
264698_at |
AT1G70200
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.241 |
-0.042 |
| 61 |
245906_at |
AT5G11070
|
unknown |
0.239 |
-0.004 |
| 62 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
0.237 |
0.021 |
| 63 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.237 |
0.064 |
| 64 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
0.237 |
0.085 |
| 65 |
251676_at |
AT3G57320
|
unknown |
0.235 |
-0.053 |
| 66 |
245130_at |
AT2G45340
|
[Leucine-rich repeat protein kinase family protein] |
0.235 |
-0.052 |
| 67 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
0.233 |
-0.001 |
| 68 |
245699_at |
AT5G04250
|
[Cysteine proteinases superfamily protein] |
0.233 |
0.007 |
| 69 |
265057_at |
AT1G52140
|
unknown |
0.232 |
0.035 |
| 70 |
250751_at |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
0.227 |
-0.119 |
| 71 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
0.226 |
-0.020 |
| 72 |
251059_at |
AT5G01810
|
ATPK10, PROTEIN KINASE 10, CIPK15, CBL-interacting protein kinase 15, PKS3, SIP2, SOS3-INTERACTING PROTEIN 2, SNRK3.1, SNF1-RELATED PROTEIN KINASE 3.1 |
0.225 |
0.076 |
| 73 |
252851_at |
AT4G40080
|
[ENTH/ANTH/VHS superfamily protein] |
0.223 |
-0.003 |
| 74 |
260126_at |
AT1G36370
|
SHM7, serine hydroxymethyltransferase 7 |
0.222 |
0.013 |
| 75 |
252427_at |
AT3G47640
|
PYE, POPEYE |
0.211 |
-0.000 |
| 76 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.208 |
0.006 |
| 77 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
0.208 |
0.009 |
| 78 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
0.207 |
-0.010 |
| 79 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.144 |
0.489 |
| 80 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-1.048 |
0.499 |
| 81 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
-1.007 |
0.379 |
| 82 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.936 |
0.360 |
| 83 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
-0.915 |
0.353 |
| 84 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
-0.888 |
0.444 |
| 85 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
-0.880 |
0.207 |
| 86 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
-0.870 |
0.328 |
| 87 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
-0.863 |
0.210 |
| 88 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-0.806 |
0.166 |
| 89 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.793 |
0.323 |
| 90 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.786 |
0.233 |
| 91 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
-0.778 |
0.188 |
| 92 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
-0.761 |
0.291 |
| 93 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
-0.753 |
0.305 |
| 94 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.747 |
0.106 |
| 95 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.740 |
0.317 |
| 96 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
-0.726 |
0.349 |
| 97 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.724 |
0.201 |
| 98 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
-0.722 |
0.306 |
| 99 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
-0.718 |
0.306 |
| 100 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.705 |
0.499 |
| 101 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
-0.698 |
0.281 |
| 102 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
-0.696 |
0.277 |
| 103 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.688 |
0.359 |
| 104 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-0.686 |
0.120 |
| 105 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
-0.673 |
0.376 |
| 106 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
-0.672 |
0.244 |
| 107 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
-0.670 |
0.230 |
| 108 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
-0.639 |
0.195 |
| 109 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.639 |
0.146 |
| 110 |
254229_at |
AT4G23610
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.624 |
0.207 |
| 111 |
257101_at |
AT3G25020
|
AtRLP42, receptor like protein 42, RBPG1, response to Botrytis polygalacturonases 1, RLP42, receptor like protein 42 |
-0.608 |
0.087 |
| 112 |
259489_at |
AT1G15790
|
unknown |
-0.602 |
0.262 |
| 113 |
255319_at |
AT4G04220
|
AtRLP46, receptor like protein 46, RLP46, receptor like protein 46 |
-0.595 |
0.111 |
| 114 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
-0.588 |
0.268 |
| 115 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-0.586 |
0.174 |
| 116 |
252346_at |
AT3G48650
|
[pseudogene] |
-0.581 |
0.237 |
| 117 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
-0.578 |
0.335 |
| 118 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
-0.574 |
0.085 |
| 119 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.563 |
0.079 |
| 120 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
-0.561 |
0.356 |
| 121 |
251422_at |
AT3G60540
|
[Preprotein translocase Sec, Sec61-beta subunit protein] |
-0.556 |
0.233 |
| 122 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.524 |
0.394 |
| 123 |
263202_at |
AT1G05630
|
5PTASE13, inositol-polyphosphate 5-phosphatase 13, AT5PTASE13, Arabidopsis thaliana inositol-polyphosphate 5-phosphatase 13 |
-0.519 |
0.014 |
| 124 |
259852_at |
AT1G72280
|
AERO1, endoplasmic reticulum oxidoreductins 1, ERO1, endoplasmic reticulum oxidoreductins 1 |
-0.505 |
0.251 |
| 125 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.504 |
0.145 |
| 126 |
260648_at |
AT1G08050
|
[Zinc finger (C3HC4-type RING finger) family protein] |
-0.503 |
0.196 |
| 127 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
-0.499 |
0.127 |
| 128 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
-0.487 |
0.055 |
| 129 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.484 |
0.264 |
| 130 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
-0.484 |
0.137 |
| 131 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
-0.483 |
0.222 |
| 132 |
248889_at |
AT5G46230
|
unknown |
-0.480 |
0.101 |
| 133 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.470 |
0.170 |
| 134 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
-0.467 |
0.179 |
| 135 |
259065_at |
AT3G07520
|
ATGLR1.4, GLR1.4, glutamate receptor 1.4 |
-0.462 |
0.152 |
| 136 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.456 |
0.118 |
| 137 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
-0.454 |
0.203 |
| 138 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.448 |
0.301 |
| 139 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
-0.443 |
0.069 |
| 140 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
-0.440 |
0.096 |
| 141 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
-0.437 |
0.216 |
| 142 |
258787_at |
AT3G11840
|
PUB24, plant U-box 24 |
-0.435 |
0.127 |
| 143 |
252862_at |
AT4G39830
|
[Cupredoxin superfamily protein] |
-0.427 |
0.179 |
| 144 |
247740_at |
AT5G58940
|
CRCK1, calmodulin-binding receptor-like cytoplasmic kinase 1 |
-0.425 |
0.269 |
| 145 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
-0.419 |
0.391 |
| 146 |
255121_at |
AT4G08480
|
MAPKKK9, mitogen-activated protein kinase kinase kinase 9, MEKK2, MAPK/ERK KINASE KINASE 2, SUMM1, suppressor of mkk1 mkk2 1 |
-0.410 |
0.110 |
| 147 |
258203_at |
AT3G13950
|
unknown |
-0.408 |
0.256 |
| 148 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
-0.405 |
0.231 |
| 149 |
267016_at |
AT2G39160
|
unknown |
-0.397 |
0.026 |
| 150 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.394 |
0.050 |
| 151 |
251840_at |
AT3G54960
|
ATPDI1, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 1, ATPDIL1-3, PDI-like 1-3, PDI1, PROTEIN DISULFIDE ISOMERASE 1, PDIL1-3, PDI-like 1-3 |
-0.393 |
0.089 |
| 152 |
254190_at |
AT4G23885
|
unknown |
-0.386 |
0.102 |
| 153 |
251232_at |
AT3G62780
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.380 |
-0.002 |
| 154 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
-0.380 |
0.076 |
| 155 |
259757_at |
AT1G77510
|
ATPDI6, PROTEIN DISULFIDE ISOMERASE 6, ATPDIL1-2, PDI-like 1-2, PDI6, PROTEIN DISULFIDE ISOMERASE 6, PDIL1-2, PDI-like 1-2 |
-0.378 |
0.123 |