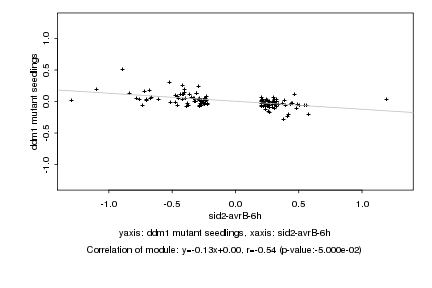
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
sid2-avrB-6h |
Log2 signal ratio
ddm1 mutant seedlings |
| 1 |
252014_at |
AT3G52870
|
[IQ calmodulin-binding motif family protein] |
1.188 |
0.043 |
| 2 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.570 |
-0.197 |
| 3 |
265387_at |
AT2G20670
|
unknown |
0.558 |
-0.054 |
| 4 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.541 |
-0.051 |
| 5 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.502 |
-0.057 |
| 6 |
245755_at |
AT1G35210
|
unknown |
0.483 |
-0.032 |
| 7 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.478 |
-0.097 |
| 8 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.461 |
0.118 |
| 9 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
0.448 |
-0.019 |
| 10 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
0.438 |
-0.022 |
| 11 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
0.433 |
-0.043 |
| 12 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
0.418 |
-0.200 |
| 13 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.404 |
-0.228 |
| 14 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.391 |
-0.050 |
| 15 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
0.383 |
0.028 |
| 16 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.376 |
-0.285 |
| 17 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
0.371 |
-0.028 |
| 18 |
247913_at |
AT5G57510
|
unknown |
0.339 |
-0.042 |
| 19 |
253819_at |
AT4G28350
|
LecRK-VII.2, L-type lectin receptor kinase VII.2 |
0.324 |
-0.065 |
| 20 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.318 |
0.042 |
| 21 |
248959_at |
AT5G45630
|
unknown |
0.317 |
0.012 |
| 22 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.317 |
0.004 |
| 23 |
254905_at |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
0.314 |
-0.022 |
| 24 |
247693_at |
AT5G59730
|
ATEXO70H7, exocyst subunit exo70 family protein H7, EXO70H7, exocyst subunit exo70 family protein H7 |
0.313 |
-0.044 |
| 25 |
255884_at |
AT1G20310
|
unknown |
0.305 |
0.039 |
| 26 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.305 |
0.013 |
| 27 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.301 |
-0.035 |
| 28 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.301 |
-0.104 |
| 29 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.300 |
0.068 |
| 30 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.293 |
0.005 |
| 31 |
263799_at |
AT2G24550
|
unknown |
0.290 |
-0.009 |
| 32 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.287 |
-0.080 |
| 33 |
251281_at |
AT3G61640
|
AGP20, arabinogalactan protein 20, AtAGP20, Arabinogalactan protein 20 |
0.287 |
0.000 |
| 34 |
261220_at |
AT1G19970
|
[ER lumen protein retaining receptor family protein] |
0.286 |
-0.032 |
| 35 |
266486_at |
AT2G47950
|
unknown |
0.267 |
0.006 |
| 36 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.267 |
-0.059 |
| 37 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
0.266 |
-0.085 |
| 38 |
246200_at |
AT4G37240
|
unknown |
0.266 |
-0.164 |
| 39 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.263 |
-0.056 |
| 40 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
0.260 |
-0.029 |
| 41 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
0.256 |
-0.154 |
| 42 |
261455_at |
AT1G21070
|
[Nucleotide-sugar transporter family protein] |
0.253 |
-0.078 |
| 43 |
263182_at |
AT1G05575
|
unknown |
0.246 |
-0.032 |
| 44 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.246 |
0.028 |
| 45 |
254408_at |
AT4G21390
|
B120 |
0.243 |
0.017 |
| 46 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.243 |
0.037 |
| 47 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.236 |
-0.076 |
| 48 |
253919_at |
AT4G27350
|
unknown |
0.233 |
-0.024 |
| 49 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.231 |
-0.115 |
| 50 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.230 |
-0.018 |
| 51 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.227 |
-0.062 |
| 52 |
257401_at |
AT1G23550
|
SRO2, similar to RCD one 2 |
0.227 |
-0.018 |
| 53 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
0.225 |
-0.051 |
| 54 |
250149_at |
AT5G14700
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.217 |
0.009 |
| 55 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.216 |
-0.064 |
| 56 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.214 |
0.000 |
| 57 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.212 |
0.033 |
| 58 |
250449_at |
AT5G10830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.210 |
0.034 |
| 59 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.204 |
0.015 |
| 60 |
247707_at |
AT5G59450
|
[GRAS family transcription factor] |
0.201 |
0.017 |
| 61 |
247597_at |
AT5G60860
|
AtRABA1f, RAB GTPase homolog A1F, RABA1f, RAB GTPase homolog A1F |
0.200 |
-0.020 |
| 62 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.199 |
0.070 |
| 63 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.199 |
-0.052 |
| 64 |
254063_at |
AT4G25390
|
[Protein kinase superfamily protein] |
0.198 |
-0.066 |
| 65 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-1.301 |
0.029 |
| 66 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.098 |
0.196 |
| 67 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-0.899 |
0.518 |
| 68 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.843 |
0.131 |
| 69 |
264958_at |
AT1G76960
|
unknown |
-0.789 |
0.052 |
| 70 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.764 |
0.037 |
| 71 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-0.739 |
-0.057 |
| 72 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.719 |
0.170 |
| 73 |
249743_at |
AT5G24540
|
BGLU31, beta glucosidase 31 |
-0.710 |
0.037 |
| 74 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.710 |
0.032 |
| 75 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
-0.680 |
0.187 |
| 76 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.678 |
0.059 |
| 77 |
261221_at |
AT1G19960
|
unknown |
-0.670 |
0.078 |
| 78 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.610 |
0.039 |
| 79 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.522 |
0.307 |
| 80 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
-0.518 |
-0.001 |
| 81 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.481 |
-0.007 |
| 82 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
-0.477 |
0.107 |
| 83 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
-0.462 |
0.091 |
| 84 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
-0.462 |
-0.053 |
| 85 |
259489_at |
AT1G15790
|
unknown |
-0.457 |
0.063 |
| 86 |
248333_at |
AT5G52390
|
[PAR1 protein] |
-0.437 |
0.127 |
| 87 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
-0.425 |
0.121 |
| 88 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.424 |
0.266 |
| 89 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-0.420 |
0.033 |
| 90 |
251419_at |
AT3G60470
|
unknown |
-0.415 |
0.127 |
| 91 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.409 |
0.205 |
| 92 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.403 |
0.147 |
| 93 |
245076_at |
AT2G23170
|
GH3.3 |
-0.397 |
0.060 |
| 94 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.394 |
-0.074 |
| 95 |
250810_at |
AT5G05090
|
[Homeodomain-like superfamily protein] |
-0.380 |
-0.027 |
| 96 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
-0.372 |
-0.058 |
| 97 |
245611_at |
AT4G14390
|
[Ankyrin repeat family protein] |
-0.368 |
0.121 |
| 98 |
257591_at |
AT3G24900
|
AtRLP39, receptor like protein 39, RLP39, receptor like protein 39 |
-0.354 |
0.069 |
| 99 |
260884_at |
AT1G29240
|
unknown |
-0.334 |
0.052 |
| 100 |
267412_at |
AT2G34940
|
BP80-3;2, binding protein of 80 kDa 3;2, VSR3;2, VACUOLAR SORTING RECEPTOR 3;2, VSR5, VACUOLAR SORTING RECEPTOR 5 |
-0.329 |
0.005 |
| 101 |
262888_at |
AT1G14790
|
AtRDR1, ATRDRP1, RDR1, RNA-dependent RNA polymerase 1 |
-0.327 |
0.052 |
| 102 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.319 |
0.006 |
| 103 |
264998_at |
AT1G67330
|
unknown |
-0.316 |
0.006 |
| 104 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
-0.310 |
0.130 |
| 105 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
-0.309 |
0.137 |
| 106 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.298 |
0.252 |
| 107 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
-0.293 |
0.046 |
| 108 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.288 |
-0.065 |
| 109 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
-0.288 |
0.022 |
| 110 |
255627_at |
AT4G00955
|
unknown |
-0.288 |
-0.052 |
| 111 |
251232_at |
AT3G62780
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.285 |
0.049 |
| 112 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
-0.282 |
-0.027 |
| 113 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.274 |
-0.057 |
| 114 |
248908_at |
AT5G45800
|
MEE62, maternal effect embryo arrest 62 |
-0.272 |
0.019 |
| 115 |
265189_at |
AT1G23840
|
unknown |
-0.272 |
0.016 |
| 116 |
257745_at |
AT3G29240
|
unknown |
-0.261 |
-0.013 |
| 117 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
-0.255 |
0.022 |
| 118 |
259472_at |
AT1G18910
|
[zinc ion binding] |
-0.247 |
-0.035 |
| 119 |
256962_at |
AT3G13560
|
[O-Glycosyl hydrolases family 17 protein] |
-0.247 |
0.053 |
| 120 |
261481_at |
AT1G14260
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.246 |
-0.024 |
| 121 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
-0.242 |
-0.031 |
| 122 |
258942_at |
AT3G09960
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.241 |
0.049 |
| 123 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
-0.237 |
0.028 |
| 124 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
-0.231 |
0.026 |
| 125 |
253238_at |
AT4G34480
|
[O-Glycosyl hydrolases family 17 protein] |
-0.229 |
0.082 |
| 126 |
262768_at |
AT1G12990
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
-0.227 |
-0.043 |
| 127 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.225 |
-0.022 |