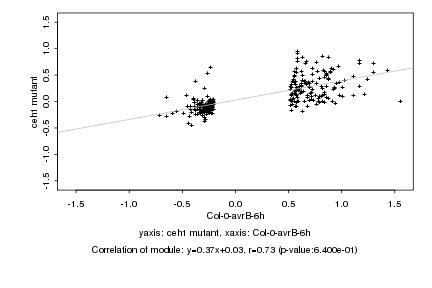
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0-avrB-6h |
Log2 signal ratio
ceh1 mutant |
| 1 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.550 |
0.015 |
| 2 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
1.425 |
0.592 |
| 3 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
1.296 |
0.726 |
| 4 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
1.293 |
0.561 |
| 5 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
1.241 |
0.428 |
| 6 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
1.210 |
0.148 |
| 7 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
1.166 |
0.291 |
| 8 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
1.163 |
0.793 |
| 9 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
1.161 |
0.737 |
| 10 |
249004_at |
AT5G44570
|
unknown |
1.111 |
0.122 |
| 11 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.107 |
0.480 |
| 12 |
252511_at |
AT3G46280
|
[protein kinase-related] |
1.019 |
0.408 |
| 13 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.003 |
0.282 |
| 14 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
1.001 |
0.105 |
| 15 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.977 |
0.126 |
| 16 |
254408_at |
AT4G21390
|
B120 |
0.974 |
0.371 |
| 17 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.969 |
0.665 |
| 18 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.945 |
0.342 |
| 19 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
0.933 |
-0.033 |
| 20 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.926 |
0.264 |
| 21 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.924 |
0.279 |
| 22 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.916 |
0.241 |
| 23 |
249896_at |
AT5G22530
|
unknown |
0.914 |
0.622 |
| 24 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.913 |
0.003 |
| 25 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.895 |
0.626 |
| 26 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.894 |
0.549 |
| 27 |
260744_at |
AT1G15010
|
unknown |
0.888 |
0.585 |
| 28 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.874 |
0.430 |
| 29 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.871 |
0.844 |
| 30 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
0.868 |
0.451 |
| 31 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.868 |
0.064 |
| 32 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.855 |
0.532 |
| 33 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.853 |
0.502 |
| 34 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.852 |
0.292 |
| 35 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.850 |
0.091 |
| 36 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.848 |
0.519 |
| 37 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.837 |
0.180 |
| 38 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.837 |
0.455 |
| 39 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.836 |
0.146 |
| 40 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.835 |
-0.003 |
| 41 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.835 |
0.574 |
| 42 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.826 |
0.599 |
| 43 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.817 |
0.130 |
| 44 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.816 |
0.861 |
| 45 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
0.815 |
0.002 |
| 46 |
249743_at |
AT5G24540
|
BGLU31, beta glucosidase 31 |
0.815 |
0.261 |
| 47 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.808 |
0.113 |
| 48 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
0.807 |
0.285 |
| 49 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.787 |
-0.017 |
| 50 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.782 |
0.442 |
| 51 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.782 |
0.096 |
| 52 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.780 |
0.014 |
| 53 |
266658_at |
AT2G25735
|
unknown |
0.774 |
0.065 |
| 54 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.764 |
0.570 |
| 55 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.757 |
0.739 |
| 56 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.756 |
-0.056 |
| 57 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.754 |
0.353 |
| 58 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
0.753 |
0.119 |
| 59 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
0.731 |
0.037 |
| 60 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.725 |
0.524 |
| 61 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.721 |
0.626 |
| 62 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.720 |
0.245 |
| 63 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.716 |
0.397 |
| 64 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.715 |
0.099 |
| 65 |
246366_at |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
0.712 |
0.053 |
| 66 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.709 |
0.176 |
| 67 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.701 |
0.170 |
| 68 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
0.697 |
0.026 |
| 69 |
264580_at |
AT1G05340
|
unknown |
0.696 |
0.279 |
| 70 |
264757_at |
AT1G61360
|
[S-locus lectin protein kinase family protein] |
0.694 |
0.290 |
| 71 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.689 |
0.344 |
| 72 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.688 |
0.301 |
| 73 |
256177_at |
AT1G51620
|
[Protein kinase superfamily protein] |
0.686 |
0.373 |
| 74 |
245089_at |
AT2G45290
|
TKL2, transketolase 2 |
0.673 |
0.082 |
| 75 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.671 |
0.117 |
| 76 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.670 |
-0.083 |
| 77 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.667 |
0.767 |
| 78 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.664 |
0.330 |
| 79 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.659 |
0.735 |
| 80 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.659 |
0.343 |
| 81 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.654 |
0.383 |
| 82 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
0.647 |
0.015 |
| 83 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.645 |
0.389 |
| 84 |
246406_at |
AT1G57650
|
[ATP binding] |
0.640 |
0.203 |
| 85 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.629 |
0.415 |
| 86 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.627 |
0.839 |
| 87 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.626 |
-0.174 |
| 88 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.624 |
0.508 |
| 89 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.623 |
0.400 |
| 90 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.618 |
0.175 |
| 91 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.617 |
0.290 |
| 92 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.615 |
0.203 |
| 93 |
251513_at |
AT3G59220
|
ATPIRIN1, PRN, pirin, PRN1 |
0.614 |
0.308 |
| 94 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
0.613 |
0.344 |
| 95 |
258203_at |
AT3G13950
|
unknown |
0.613 |
0.573 |
| 96 |
257517_at |
AT3G16330
|
unknown |
0.603 |
0.154 |
| 97 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.600 |
0.289 |
| 98 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.598 |
0.256 |
| 99 |
259033_at |
AT3G09405
|
[Pectinacetylesterase family protein] |
0.594 |
0.299 |
| 100 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
0.592 |
0.222 |
| 101 |
252604_at |
AT3G45060
|
ATNRT2.6, ARABIDOPSIS THALIANA HIGH AFFINITY NITRATE TRANSPORTER 2.6, NRT2.6, high affinity nitrate transporter 2.6 |
0.589 |
0.012 |
| 102 |
256627_at |
AT3G19970
|
[alpha/beta-Hydrolases superfamily protein] |
0.586 |
0.240 |
| 103 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.584 |
0.018 |
| 104 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.582 |
0.950 |
| 105 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.579 |
0.923 |
| 106 |
260754_at |
AT1G49000
|
unknown |
0.576 |
0.087 |
| 107 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
0.576 |
0.829 |
| 108 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.575 |
0.757 |
| 109 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.574 |
0.277 |
| 110 |
265188_at |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
0.574 |
-0.085 |
| 111 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
0.572 |
0.116 |
| 112 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
0.571 |
0.632 |
| 113 |
257058_at |
AT3G15352
|
ATCOX17, ARABIDOPSIS THALIANA CYTOCHROME C OXIDASE 17, COX17, cytochrome c oxidase 17 |
0.569 |
0.215 |
| 114 |
265648_at |
AT2G27500
|
[Glycosyl hydrolase superfamily protein] |
0.568 |
0.000 |
| 115 |
263221_at |
AT1G30620
|
HSR8, HIGH SUGAR RESPONSE8, MUR4, MURUS 4, UXE1, UDP-D-XYLOSE 4-EPIMERASE 1 |
0.567 |
0.133 |
| 116 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.566 |
0.566 |
| 117 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
0.564 |
-0.083 |
| 118 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.560 |
0.190 |
| 119 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.558 |
0.339 |
| 120 |
265276_at |
AT2G28400
|
unknown |
0.558 |
0.212 |
| 121 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.556 |
0.393 |
| 122 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.552 |
0.164 |
| 123 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
0.550 |
0.369 |
| 124 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.549 |
0.012 |
| 125 |
251633_at |
AT3G57460
|
[catalytics] |
0.549 |
0.486 |
| 126 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.548 |
0.582 |
| 127 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.547 |
0.499 |
| 128 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.543 |
0.070 |
| 129 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.541 |
0.428 |
| 130 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.534 |
0.052 |
| 131 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
0.533 |
0.168 |
| 132 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.529 |
0.384 |
| 133 |
251984_at |
AT3G53260
|
ATPAL2, PAL2, phenylalanine ammonia-lyase 2 |
0.528 |
-0.055 |
| 134 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
0.527 |
-0.153 |
| 135 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.525 |
0.266 |
| 136 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
0.525 |
0.012 |
| 137 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.523 |
0.143 |
| 138 |
247532_at |
AT5G61560
|
[U-box domain-containing protein kinase family protein] |
0.522 |
0.118 |
| 139 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.522 |
0.331 |
| 140 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.518 |
-0.068 |
| 141 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.514 |
0.232 |
| 142 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
0.514 |
0.027 |
| 143 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.513 |
0.057 |
| 144 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.512 |
0.295 |
| 145 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.719 |
-0.247 |
| 146 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.657 |
-0.271 |
| 147 |
256569_at |
AT3G19550
|
unknown |
-0.655 |
0.085 |
| 148 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.595 |
-0.208 |
| 149 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.556 |
-0.182 |
| 150 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.493 |
-0.215 |
| 151 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
-0.465 |
0.121 |
| 152 |
262753_at |
AT1G16340
|
ATKDSA2, ATKSDA |
-0.456 |
-0.079 |
| 153 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.448 |
-0.413 |
| 154 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.441 |
-0.270 |
| 155 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.428 |
-0.093 |
| 156 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
-0.427 |
-0.132 |
| 157 |
252215_at |
AT3G50240
|
KICP-02 |
-0.422 |
-0.190 |
| 158 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.418 |
-0.437 |
| 159 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.404 |
0.070 |
| 160 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.396 |
0.040 |
| 161 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
-0.394 |
-0.004 |
| 162 |
247288_at |
AT5G64330
|
JK218, NPH3, NON-PHOTOTROPIC HYPOCOTYL 3, RPT3, ROOT PHOTOTROPISM 3 |
-0.388 |
-0.093 |
| 163 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.387 |
-0.141 |
| 164 |
258109_at |
AT3G23640
|
HGL1, heteroglycan glucosidase 1 |
-0.387 |
-0.003 |
| 165 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.382 |
-0.169 |
| 166 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.379 |
-0.234 |
| 167 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-0.378 |
0.388 |
| 168 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.363 |
-0.141 |
| 169 |
248420_at |
AT5G51560
|
[Leucine-rich repeat protein kinase family protein] |
-0.363 |
-0.047 |
| 170 |
246231_at |
AT4G37080
|
unknown |
-0.361 |
-0.233 |
| 171 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.360 |
0.033 |
| 172 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
-0.360 |
-0.110 |
| 173 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.345 |
-0.099 |
| 174 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
-0.341 |
-0.042 |
| 175 |
264916_at |
AT1G60810
|
ACLA-2, ATP-citrate lyase A-2 |
-0.341 |
-0.021 |
| 176 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.339 |
-0.196 |
| 177 |
257236_at |
AT3G15095
|
HCF243, high chlorophyll fluorescence 243 |
-0.339 |
-0.087 |
| 178 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.339 |
-0.145 |
| 179 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.338 |
-0.225 |
| 180 |
245348_at |
AT4G17770
|
ATTPS5, trehalose phosphatase/synthase 5, TPS5, TREHALOSE -6-PHOSPHATASE SYNTHASE S5, TPS5, trehalose phosphatase/synthase 5 |
-0.336 |
-0.108 |
| 181 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.335 |
-0.158 |
| 182 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.334 |
-0.197 |
| 183 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.331 |
-0.074 |
| 184 |
253920_at |
AT4G27230
|
HTA2, histone H2A 2 |
-0.324 |
-0.065 |
| 185 |
248613_at |
AT5G49555
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.324 |
-0.061 |
| 186 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.322 |
-0.197 |
| 187 |
260902_at |
AT1G21440
|
[Phosphoenolpyruvate carboxylase family protein] |
-0.321 |
-0.014 |
| 188 |
264930_at |
AT1G60800
|
AtNIK3, NIK3, NSP-interacting kinase 3 |
-0.318 |
-0.134 |
| 189 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.317 |
-0.251 |
| 190 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.317 |
-0.184 |
| 191 |
245891_at |
AT5G09220
|
AAP2, amino acid permease 2 |
-0.316 |
0.020 |
| 192 |
255876_at |
AT2G40480
|
unknown |
-0.315 |
-0.062 |
| 193 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
-0.314 |
-0.167 |
| 194 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.311 |
0.031 |
| 195 |
256400_at |
AT3G06140
|
LUL4, LOG2-LIKE UBIQUITIN LIGASE4 |
-0.311 |
-0.160 |
| 196 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.310 |
-0.137 |
| 197 |
245845_at |
AT1G26150
|
AtPERK10, proline-rich extensin-like receptor kinase 10, PERK10, proline-rich extensin-like receptor kinase 10 |
-0.310 |
-0.202 |
| 198 |
256962_at |
AT3G13560
|
[O-Glycosyl hydrolases family 17 protein] |
-0.308 |
-0.063 |
| 199 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
-0.305 |
-0.179 |
| 200 |
266054_at |
AT2G40640
|
[RING/U-box superfamily protein] |
-0.305 |
-0.080 |
| 201 |
251330_at |
AT3G61550
|
[RING/U-box superfamily protein] |
-0.305 |
-0.164 |
| 202 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.302 |
-0.086 |
| 203 |
265726_at |
AT2G32010
|
CVL1, CVP2 like 1 |
-0.301 |
-0.167 |
| 204 |
259996_at |
AT1G67910
|
unknown |
-0.297 |
-0.073 |
| 205 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.296 |
-0.364 |
| 206 |
264958_at |
AT1G76960
|
unknown |
-0.295 |
0.260 |
| 207 |
263696_at |
AT1G31230
|
AK-HSDH, ASPARTATE KINASE-HOMOSERINE DEHYDROGENASE, AK-HSDH I, aspartate kinase-homoserine dehydrogenase i |
-0.294 |
-0.082 |
| 208 |
252033_at |
AT3G51950
|
[Zinc finger (CCCH-type) family protein / RNA recognition motif (RRM)-containing protein] |
-0.293 |
-0.107 |
| 209 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.292 |
-0.192 |
| 210 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.292 |
-0.290 |
| 211 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.289 |
-0.333 |
| 212 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-0.288 |
-0.042 |
| 213 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.283 |
-0.099 |
| 214 |
245592_at |
AT4G14540
|
NF-YB3, nuclear factor Y, subunit B3 |
-0.280 |
-0.134 |
| 215 |
262162_at |
AT1G78020
|
unknown |
-0.279 |
-0.170 |
| 216 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.279 |
-0.228 |
| 217 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.278 |
-0.047 |
| 218 |
258828_at |
AT3G07130
|
ATPAP15, PURPLE ACID PHOSPHATASE 15, PAP15, purple acid phosphatase 15 |
-0.278 |
-0.079 |
| 219 |
265673_at |
AT2G32090
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-0.275 |
-0.088 |
| 220 |
258156_at |
AT3G18050
|
unknown |
-0.275 |
-0.111 |
| 221 |
258989_at |
AT3G08920
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.274 |
-0.084 |
| 222 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.271 |
-0.168 |
| 223 |
253545_at |
AT4G31310
|
[AIG2-like (avirulence induced gene) family protein] |
-0.270 |
-0.087 |
| 224 |
261221_at |
AT1G19960
|
unknown |
-0.268 |
0.098 |
| 225 |
256872_at |
AT3G26490
|
[Phototropic-responsive NPH3 family protein] |
-0.267 |
-0.130 |
| 226 |
263442_at |
AT2G28605
|
[Photosystem II reaction center PsbP family protein] |
-0.267 |
-0.171 |
| 227 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
-0.266 |
-0.042 |
| 228 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
-0.265 |
0.532 |
| 229 |
260381_at |
AT1G73860
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.263 |
-0.097 |
| 230 |
254198_at |
AT4G24090
|
unknown |
-0.260 |
-0.127 |
| 231 |
248291_at |
AT5G53020
|
[Ribonuclease P protein subunit P38-related] |
-0.259 |
-0.123 |
| 232 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.259 |
-0.200 |
| 233 |
260462_at |
AT1G10970
|
ATZIP4, ZIP4, zinc transporter 4 precursor |
-0.259 |
-0.096 |
| 234 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.259 |
-0.047 |
| 235 |
262170_at |
AT1G74940
|
unknown |
-0.259 |
0.010 |
| 236 |
267157_at |
AT2G37630
|
AS1, ASYMMETRIC LEAVES 1, ATMYB91, MYB DOMAIN PROTEIN 91, ATPHAN, ARABIDOPSIS PHANTASTICA-LIKE 1, MYB91, MYB DOMAIN PROTEIN 91 |
-0.257 |
-0.031 |
| 237 |
264900_at |
AT1G23080
|
ATPIN7, ARABIDOPSIS PIN-FORMED 7, PIN7, PIN-FORMED 7 |
-0.257 |
-0.126 |
| 238 |
251500_at |
AT3G59110
|
[Protein kinase superfamily protein] |
-0.255 |
-0.077 |
| 239 |
248329_at |
AT5G52780
|
unknown |
-0.254 |
-0.155 |
| 240 |
262370_at |
AT1G73090
|
unknown |
-0.253 |
-0.100 |
| 241 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.252 |
-0.131 |
| 242 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.251 |
-0.226 |
| 243 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.250 |
-0.199 |
| 244 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.249 |
-0.143 |
| 245 |
266635_at |
AT2G35470
|
unknown |
-0.246 |
-0.051 |
| 246 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.245 |
0.034 |
| 247 |
246904_at |
AT5G25480
|
AtDNMT2, DNA METHYLTRANSFERASE 2, DNMT2, DNA methyltransferase-2 |
-0.244 |
-0.021 |
| 248 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
-0.244 |
0.653 |
| 249 |
253010_at |
AT4G37750
|
ANT, AINTEGUMENTA, CKC, CKC1, COMPLEMENTING A PROTEIN KINASE C MUTANT 1, DRG, DRAGON |
-0.244 |
-0.048 |
| 250 |
247093_at |
AT5G66350
|
SHI, SHORT INTERNODES |
-0.243 |
0.038 |
| 251 |
246551_at |
AT5G15070
|
[Phosphoglycerate mutase-like family protein] |
-0.243 |
-0.049 |
| 252 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.243 |
-0.003 |
| 253 |
252353_at |
AT3G48200
|
unknown |
-0.240 |
-0.135 |
| 254 |
258535_at |
AT3G06750
|
[hydroxyproline-rich glycoprotein family protein] |
-0.240 |
-0.203 |
| 255 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.238 |
-0.068 |
| 256 |
260631_at |
AT1G62350
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.236 |
-0.005 |
| 257 |
247747_at |
AT5G59000
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.236 |
-0.052 |
| 258 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
-0.235 |
-0.058 |
| 259 |
264822_at |
AT1G03457
|
AtBRN2, BRN2, Bruno-like 2 |
-0.235 |
0.049 |
| 260 |
255788_at |
AT2G33310
|
IAA13, auxin-induced protein 13 |
-0.234 |
-0.103 |
| 261 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.233 |
-0.145 |
| 262 |
262247_at |
AT1G48420
|
ACD1, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID DEAMINASE 1, ATACD1, A. THALIANA 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID DEAMINASE 1, AtDCD, D-CDES, D-cysteine desulfhydrase, DCD, D-cysteine desulfhydrase |
-0.231 |
-0.025 |
| 263 |
265998_at |
AT2G24270
|
ALDH11A3, aldehyde dehydrogenase 11A3 |
-0.231 |
-0.047 |
| 264 |
259723_at |
AT1G60960
|
ATIRT3, IRON REGULATED TRANSPORTER 3, IRT3, iron regulated transporter 3 |
-0.229 |
-0.212 |
| 265 |
260106_at |
AT1G35420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.229 |
-0.078 |
| 266 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.228 |
0.016 |
| 267 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.227 |
-0.133 |
| 268 |
250582_at |
AT5G07580
|
[Integrase-type DNA-binding superfamily protein] |
-0.224 |
0.016 |
| 269 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
-0.223 |
-0.036 |
| 270 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.221 |
-0.053 |
| 271 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
-0.221 |
-0.141 |
| 272 |
266001_at |
AT2G24150
|
HHP3, heptahelical protein 3 |
-0.220 |
-0.095 |
| 273 |
256293_at |
AT1G69440
|
AGO7, ARGONAUTE7, ZIP, ZIPPY |
-0.220 |
-0.100 |
| 274 |
264865_at |
AT1G24120
|
ARL1, ARG1-like 1 |
-0.219 |
-0.076 |
| 275 |
266751_at |
AT2G47020
|
[Peptide chain release factor 1] |
-0.218 |
0.028 |
| 276 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
-0.218 |
-0.013 |
| 277 |
259017_at |
AT3G07310
|
unknown |
-0.217 |
-0.106 |
| 278 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.216 |
-0.038 |
| 279 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.216 |
-0.222 |
| 280 |
263700_at |
AT1G31150
|
[FUNCTIONS IN: sequence-specific DNA binding transcription factor activity] |
-0.216 |
-0.061 |
| 281 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
-0.216 |
0.021 |
| 282 |
258708_at |
AT3G09580
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.215 |
-0.135 |
| 283 |
249231_at |
AT5G42030
|
ABIL4, ABL interactor-like protein 4 |
-0.215 |
-0.145 |
| 284 |
262727_at |
AT1G75800
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.215 |
0.009 |
| 285 |
264590_at |
AT2G17710
|
unknown |
-0.214 |
-0.009 |
| 286 |
257132_at |
AT3G20230
|
[Ribosomal L18p/L5e family protein] |
-0.213 |
-0.041 |
| 287 |
267400_at |
AT2G26240
|
[Transmembrane proteins 14C] |
-0.212 |
0.057 |