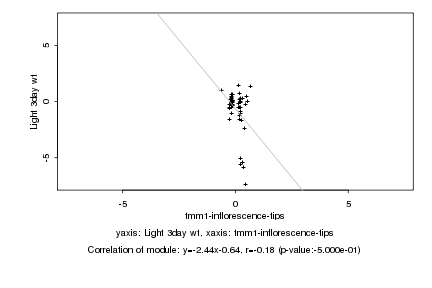
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
tmm1-inflorescence-tips |
Log2 signal ratio
Light 3day wt |
| 1 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.636 |
1.366 |
| 2 |
262040_at |
AT1G80080
|
AtRLP17, Receptor Like Protein 17, TMM, TOO MANY MOUTHS |
0.492 |
0.057 |
| 3 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.461 |
0.524 |
| 4 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.418 |
-0.190 |
| 5 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.408 |
-7.301 |
| 6 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.398 |
-2.391 |
| 7 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.357 |
-5.802 |
| 8 |
256617_at |
AT3G22240
|
unknown |
0.280 |
0.270 |
| 9 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
0.271 |
-5.395 |
| 10 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.226 |
-1.658 |
| 11 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.224 |
-5.592 |
| 12 |
254962_at |
AT4G11050
|
AtGH9C3, glycosyl hydrolase 9C3, GH9C3, glycosyl hydrolase 9C3 |
0.212 |
-1.055 |
| 13 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
0.205 |
-0.064 |
| 14 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
0.195 |
0.321 |
| 15 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.194 |
-0.820 |
| 16 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
0.188 |
-0.449 |
| 17 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.187 |
-5.051 |
| 18 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.182 |
0.028 |
| 19 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.165 |
-0.515 |
| 20 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
0.149 |
0.256 |
| 21 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.149 |
-1.531 |
| 22 |
250201_at |
AT5G14230
|
unknown |
0.147 |
-1.176 |
| 23 |
256599_at |
AT3G14760
|
unknown |
0.142 |
0.729 |
| 24 |
248819_at |
AT5G47050
|
[SBP (S-ribonuclease binding protein) family protein] |
0.133 |
-0.145 |
| 25 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
0.133 |
1.476 |
| 26 |
247906_at |
AT5G57420
|
IAA33, indole-3-acetic acid inducible 33 |
0.130 |
-0.515 |
| 27 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.633 |
1.045 |
| 28 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.289 |
-0.445 |
| 29 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
-0.288 |
-1.512 |
| 30 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
-0.286 |
-0.232 |
| 31 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.270 |
-0.533 |
| 32 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.255 |
-0.241 |
| 33 |
254787_at |
AT4G12690
|
unknown |
-0.236 |
0.264 |
| 34 |
257670_at |
AT3G20340
|
unknown |
-0.214 |
0.015 |
| 35 |
265481_at |
AT2G15960
|
unknown |
-0.213 |
0.280 |
| 36 |
250582_at |
AT5G07580
|
[Integrase-type DNA-binding superfamily protein] |
-0.206 |
0.386 |
| 37 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.205 |
-0.260 |
| 38 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
-0.202 |
0.706 |
| 39 |
259982_at |
AT1G76410
|
ATL8 |
-0.201 |
-0.993 |
| 40 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
-0.195 |
0.519 |
| 41 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.192 |
0.200 |
| 42 |
255937_at |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
-0.188 |
-0.515 |
| 43 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
-0.187 |
-0.047 |
| 44 |
253064_at |
AT4G37730
|
AtbZIP7, basic leucine-zipper 7, bZIP7, basic leucine-zipper 7 |
-0.171 |
0.046 |
| 45 |
263623_at |
AT2G04860
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.169 |
-0.246 |
| 46 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-0.168 |
0.124 |
| 47 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
-0.163 |
-0.266 |
| 48 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
-0.161 |
-0.054 |
| 49 |
258383_at |
AT3G15440
|
unknown |
-0.155 |
0.689 |
| 50 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
-0.152 |
-0.271 |
| 51 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.151 |
0.132 |
| 52 |
260640_at |
AT1G53350
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.148 |
0.095 |