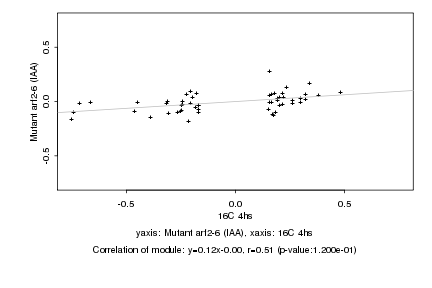
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
16C 4hs |
Log2 signal ratio
Mutant arf2-6 (IAA) |
| 1 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
0.480 |
0.092 |
| 2 |
256924_at |
AT3G29590
|
AT5MAT |
0.378 |
0.063 |
| 3 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.337 |
0.168 |
| 4 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
0.320 |
0.019 |
| 5 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.319 |
0.066 |
| 6 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.297 |
0.033 |
| 7 |
246103_at |
AT5G28640
|
AN3, ANGUSTIFOLIA 3, ATGIF1, ARABIDOPSIS GRF1-INTERACTING FACTOR 1, GIF, GRF1-INTERACTING FACTOR, GIF1, GRF1-INTERACTING FACTOR 1 |
0.296 |
-0.001 |
| 8 |
256627_at |
AT3G19970
|
[alpha/beta-Hydrolases superfamily protein] |
0.259 |
-0.017 |
| 9 |
262321_at |
AT1G27570
|
[phosphatidylinositol 3- and 4-kinase family protein] |
0.258 |
0.013 |
| 10 |
252077_at |
AT3G51720
|
unknown |
0.234 |
0.131 |
| 11 |
259333_at |
AT3G03810
|
EDA30, embryo sac development arrest 30 |
0.218 |
0.040 |
| 12 |
258560_at |
AT3G06020
|
FAF4, FANTASTIC FOUR 4 |
0.215 |
0.078 |
| 13 |
254349_at |
AT4G22250
|
[RING/U-box superfamily protein] |
0.212 |
-0.024 |
| 14 |
254563_at |
AT4G19120
|
ERD3, early-responsive to dehydration 3 |
0.201 |
0.041 |
| 15 |
254578_at |
AT4G19410
|
[Pectinacetylesterase family protein] |
0.198 |
-0.035 |
| 16 |
263258_at |
AT1G10540
|
ATNAT8, NAT8, nucleobase-ascorbate transporter 8 |
0.191 |
0.015 |
| 17 |
245570_at |
AT4G14670
|
CLPB2, casein lytic proteinase B2 |
0.190 |
0.031 |
| 18 |
247057_at |
AT5G66770
|
[GRAS family transcription factor] |
0.182 |
-0.097 |
| 19 |
248380_at |
AT5G51820
|
ATPGMP, ARABIDOPSIS THALIANA PHOSPHOGLUCOMUTASE, PGM, phosphoglucomutase, PGM1, STF1, STARCH-FREE 1 |
0.179 |
0.076 |
| 20 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.174 |
-0.125 |
| 21 |
253323_at |
AT4G33920
|
APD5, Arabidopsis Pp2c clade D 5 |
0.166 |
-0.111 |
| 22 |
253252_at |
AT4G34740
|
ASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATPURF2, CIA1, CHLOROPLAST IMPORT APPARATUS 1, DOV1, DIFFERENTIAL DEVELOPMENT OF VASCULAR ASSOCIATED CELLS |
0.164 |
0.067 |
| 23 |
258429_at |
AT3G16620
|
ATTOC120, ARABIDOPSIS THALIANA TRANSLOCON OUTER COMPLEX PROTEIN 120, TOC120, translocon outer complex protein 120 |
0.161 |
-0.009 |
| 24 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
0.156 |
0.277 |
| 25 |
251052_at |
AT5G02470
|
DPA |
0.154 |
-0.004 |
| 26 |
263119_at |
AT1G03110
|
AtTRM82, TRM82, tRNA modification 82 |
0.154 |
0.058 |
| 27 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
0.150 |
-0.072 |
| 28 |
254574_at |
AT4G19430
|
unknown |
-0.755 |
-0.162 |
| 29 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.743 |
-0.094 |
| 30 |
252345_at |
AT3G48640
|
unknown |
-0.719 |
-0.010 |
| 31 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-0.666 |
-0.000 |
| 32 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.465 |
-0.086 |
| 33 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.451 |
-0.008 |
| 34 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-0.393 |
-0.143 |
| 35 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
-0.319 |
-0.010 |
| 36 |
251986_at |
AT3G53310
|
[AP2/B3-like transcriptional factor family protein] |
-0.316 |
0.005 |
| 37 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
-0.310 |
-0.102 |
| 38 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
-0.268 |
-0.098 |
| 39 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-0.254 |
-0.084 |
| 40 |
255148_at |
AT4G08470
|
MAPKKK10, MEKK3, MAPK/ERK kinase kinase 3 |
-0.248 |
-0.081 |
| 41 |
246789_at |
AT5G27600
|
ATLACS7, LACS7, long-chain acyl-CoA synthetase 7 |
-0.248 |
-0.031 |
| 42 |
254948_at |
AT4G11000
|
[Ankyrin repeat family protein] |
-0.247 |
-0.025 |
| 43 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.244 |
0.008 |
| 44 |
246380_at |
AT1G57750
|
CYP96A15, cytochrome P450, family 96, subfamily A, polypeptide 15, MAH1, MID-CHAIN ALKANE HYDROXYLASE 1 |
-0.227 |
0.065 |
| 45 |
261221_at |
AT1G19960
|
unknown |
-0.216 |
-0.179 |
| 46 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
-0.210 |
0.096 |
| 47 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
-0.209 |
-0.011 |
| 48 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.198 |
0.045 |
| 49 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.184 |
-0.048 |
| 50 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
-0.183 |
0.080 |
| 51 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.173 |
-0.100 |
| 52 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.171 |
-0.033 |
| 53 |
253377_at |
AT4G33300
|
ADR1-L1, ADR1-like 1 |
-0.171 |
-0.028 |
| 54 |
248210_at |
AT5G54000
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.170 |
-0.069 |