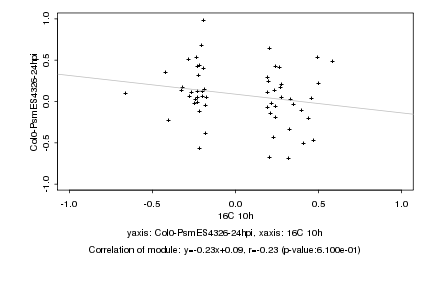
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
16C 10h |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.579 |
0.490 |
| 2 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
0.497 |
0.223 |
| 3 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.493 |
0.541 |
| 4 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.469 |
-0.468 |
| 5 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.457 |
0.044 |
| 6 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
0.435 |
-0.196 |
| 7 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.405 |
-0.502 |
| 8 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.394 |
-0.105 |
| 9 |
248614_at |
AT5G49560
|
[Putative methyltransferase family protein] |
0.349 |
-0.028 |
| 10 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.328 |
0.029 |
| 11 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
0.325 |
-0.328 |
| 12 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
0.314 |
-0.686 |
| 13 |
256518_at |
AT1G66080
|
unknown |
0.274 |
0.212 |
| 14 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.272 |
0.057 |
| 15 |
245821_at |
AT1G26270
|
[Phosphatidylinositol 3- and 4-kinase family protein] |
0.268 |
0.176 |
| 16 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.260 |
0.413 |
| 17 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
0.241 |
-0.188 |
| 18 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
0.239 |
-0.053 |
| 19 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.239 |
0.431 |
| 20 |
256320_at |
AT3G12170
|
[Chaperone DnaJ-domain superfamily protein] |
0.232 |
0.141 |
| 21 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.225 |
-0.432 |
| 22 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
0.214 |
-0.016 |
| 23 |
257237_at |
AT3G14890
|
[phosphoesterase] |
0.209 |
-0.144 |
| 24 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.202 |
-0.676 |
| 25 |
260734_at |
AT1G17600
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.201 |
0.642 |
| 26 |
260264_at |
AT1G68500
|
unknown |
0.199 |
0.245 |
| 27 |
258952_at |
AT3G01410
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.193 |
-0.064 |
| 28 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.189 |
0.293 |
| 29 |
251020_at |
AT5G02270
|
ABCI20, ATP-binding cassette I20, ATNAP9, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 9, NAP9, non-intrinsic ABC protein 9 |
0.188 |
0.116 |
| 30 |
254574_at |
AT4G19430
|
unknown |
-0.665 |
0.106 |
| 31 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
-0.422 |
0.356 |
| 32 |
262811_at |
AT1G11700
|
unknown |
-0.404 |
-0.226 |
| 33 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
-0.328 |
0.134 |
| 34 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
-0.320 |
0.178 |
| 35 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
-0.286 |
0.510 |
| 36 |
248585_at |
AT5G49640
|
unknown |
-0.278 |
0.069 |
| 37 |
257949_at |
AT3G21750
|
UGT71B1, UDP-glucosyl transferase 71B1 |
-0.266 |
0.120 |
| 38 |
251987_at |
AT3G53280
|
CYP71B5, cytochrome p450 71b5 |
-0.248 |
-0.017 |
| 39 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.244 |
0.027 |
| 40 |
257295_at |
AT3G17420
|
GPK1, glyoxysomal protein kinase 1 |
-0.235 |
0.543 |
| 41 |
260592_at |
AT1G55850
|
ATCSLE1, CSLE1, cellulose synthase like E1 |
-0.233 |
0.122 |
| 42 |
252240_at |
AT3G50010
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.231 |
0.059 |
| 43 |
264338_at |
AT1G70300
|
KUP6, K+ uptake permease 6 |
-0.230 |
0.432 |
| 44 |
248827_at |
AT5G47100
|
ATCBL9, CBL9, calcineurin B-like protein 9 |
-0.229 |
-0.011 |
| 45 |
258179_at |
AT3G21690
|
[MATE efflux family protein] |
-0.227 |
0.315 |
| 46 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.222 |
0.441 |
| 47 |
250784_at |
AT5G05480
|
[Peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A protein] |
-0.221 |
-0.120 |
| 48 |
250696_at |
AT5G06790
|
unknown |
-0.219 |
-0.567 |
| 49 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
-0.206 |
0.681 |
| 50 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.203 |
0.125 |
| 51 |
256041_at |
AT1G07230
|
NPC1, non-specific phospholipase C1 |
-0.202 |
0.062 |
| 52 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
-0.196 |
0.992 |
| 53 |
266659_at |
AT2G25850
|
PAPS2, poly(A) polymerase 2 |
-0.195 |
0.408 |
| 54 |
250754_at |
AT5G05700
|
ATATE1, ATE1, arginine-tRNA protein transferase 1, DLS1, DELAYED LEAF SENESCENCE 1 |
-0.191 |
0.157 |
| 55 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.186 |
-0.378 |
| 56 |
251391_at |
AT3G60910
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.183 |
-0.046 |
| 57 |
259032_at |
AT3G09380
|
unknown |
-0.180 |
0.049 |