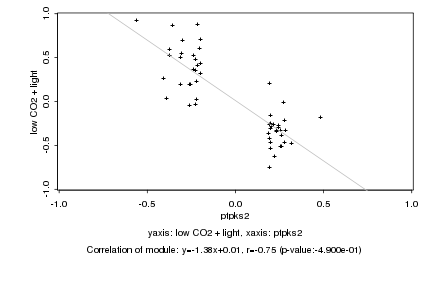
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ptpks2 |
Log2 signal ratio
low CO2 + light |
| 1 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.478 |
-0.175 |
| 2 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
0.314 |
-0.473 |
| 3 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.282 |
-0.330 |
| 4 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
0.277 |
-0.456 |
| 5 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.273 |
-0.211 |
| 6 |
249427_at |
AT5G39850
|
[Ribosomal protein S4] |
0.272 |
-0.008 |
| 7 |
265730_at |
AT2G32220
|
[Ribosomal L27e protein family] |
0.258 |
-0.503 |
| 8 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.256 |
-0.386 |
| 9 |
251638_at |
AT3G57490
|
[Ribosomal protein S5 family protein] |
0.253 |
-0.323 |
| 10 |
266510_at |
AT2G47990
|
EDA13, EMBRYO SAC DEVELOPMENT ARREST 13, EDA19, EMBRYO SAC DEVELOPMENT ARREST 19, SWA1, SLOW WALKER1 |
0.252 |
-0.508 |
| 11 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
0.243 |
-0.289 |
| 12 |
250546_at |
AT5G08180
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
0.239 |
-0.265 |
| 13 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
0.230 |
-0.341 |
| 14 |
264286_at |
AT1G61870
|
PPR336, pentatricopeptide repeat 336 |
0.230 |
-0.320 |
| 15 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.221 |
-0.623 |
| 16 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
0.211 |
-0.260 |
| 17 |
249392_at |
AT5G40150
|
[Peroxidase superfamily protein] |
0.203 |
-0.283 |
| 18 |
258474_at |
AT3G02650
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.197 |
-0.532 |
| 19 |
258155_at |
AT3G18130
|
RACK1C, receptor for activated C kinase 1C, RACK1C_AT, receptor for activated C kinase 1C |
0.197 |
-0.299 |
| 20 |
251152_at |
AT3G63130
|
ATRANGAP1, RAN GTPASE-ACTIVATING PROTEIN 1, RANGAP1, RAN GTPase activating protein 1 |
0.196 |
-0.151 |
| 21 |
245254_at |
AT4G14680
|
APS3 |
0.195 |
-0.247 |
| 22 |
253165_at |
AT4G35320
|
unknown |
0.194 |
-0.466 |
| 23 |
246762_at |
AT5G27620
|
CYCH;1, cyclin H;1 |
0.192 |
-0.255 |
| 24 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
0.191 |
0.211 |
| 25 |
252034_at |
AT3G52040
|
unknown |
0.191 |
-0.420 |
| 26 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.190 |
-0.745 |
| 27 |
266801_at |
AT2G22870
|
EMB2001, embryo defective 2001 |
0.186 |
-0.363 |
| 28 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.564 |
0.934 |
| 29 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.412 |
0.270 |
| 30 |
262719_at |
AT1G43590
|
unknown |
-0.392 |
0.043 |
| 31 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.379 |
0.529 |
| 32 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
-0.379 |
0.600 |
| 33 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.360 |
0.877 |
| 34 |
263384_at |
AT2G40130
|
SMXL8, SMAX1-like 8 |
-0.315 |
0.194 |
| 35 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
-0.312 |
0.503 |
| 36 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.307 |
0.548 |
| 37 |
265387_at |
AT2G20670
|
unknown |
-0.303 |
0.698 |
| 38 |
267110_at |
AT2G14800
|
unknown |
-0.264 |
-0.045 |
| 39 |
250766_at |
AT5G05550
|
[sequence-specific DNA binding transcription factors] |
-0.262 |
0.195 |
| 40 |
246951_at |
AT5G04880
|
[expressed protein] |
-0.255 |
0.201 |
| 41 |
254424_at |
AT4G21510
|
AtFBS2, FBS2, F-box stress induced 2 |
-0.243 |
0.376 |
| 42 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
-0.243 |
0.529 |
| 43 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.230 |
0.483 |
| 44 |
250013_at |
AT5G18040
|
unknown |
-0.229 |
-0.024 |
| 45 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.227 |
0.354 |
| 46 |
255376_x_at |
AT4G03790
|
[gypsy-like retrotransposon family (Athila), has a 2.2e-286 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-0.225 |
0.024 |
| 47 |
252036_at |
AT3G52070
|
unknown |
-0.222 |
0.236 |
| 48 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
-0.219 |
0.888 |
| 49 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-0.216 |
0.413 |
| 50 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.208 |
0.614 |
| 51 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.201 |
0.709 |
| 52 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
-0.200 |
0.434 |
| 53 |
255285_at |
AT4G04630
|
unknown |
-0.199 |
0.322 |