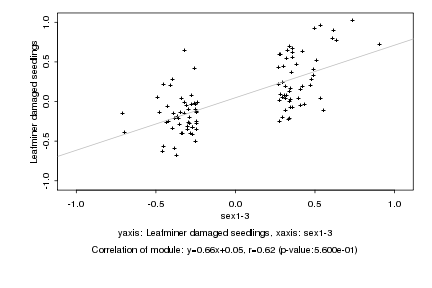
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
sex1-3 |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.902 |
0.732 |
| 2 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.730 |
1.035 |
| 3 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.629 |
0.773 |
| 4 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.616 |
0.903 |
| 5 |
258209_at |
AT3G14060
|
unknown |
0.608 |
0.801 |
| 6 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
0.553 |
-0.111 |
| 7 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.531 |
0.971 |
| 8 |
254286_at |
AT4G22950
|
AGL19, AGAMOUS-like 19, GL19 |
0.530 |
0.040 |
| 9 |
263475_at |
AT2G31945
|
unknown |
0.507 |
0.527 |
| 10 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.492 |
0.931 |
| 11 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.490 |
0.406 |
| 12 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.487 |
0.336 |
| 13 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.474 |
0.288 |
| 14 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.468 |
0.207 |
| 15 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.434 |
-0.032 |
| 16 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.420 |
0.640 |
| 17 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.417 |
0.194 |
| 18 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.406 |
0.162 |
| 19 |
258327_at |
AT3G22640
|
PAP85 |
0.405 |
-0.038 |
| 20 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.394 |
0.040 |
| 21 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.381 |
0.473 |
| 22 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.357 |
-0.070 |
| 23 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.357 |
0.627 |
| 24 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.355 |
0.559 |
| 25 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
0.354 |
0.672 |
| 26 |
247835_at |
AT5G57910
|
unknown |
0.347 |
0.378 |
| 27 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.344 |
0.168 |
| 28 |
245807_at |
AT1G46768
|
RAP2.1, related to AP2 1 |
0.343 |
-0.071 |
| 29 |
248969_at |
AT5G45310
|
unknown |
0.342 |
0.028 |
| 30 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
0.338 |
0.132 |
| 31 |
259103_at |
AT3G11690
|
unknown |
0.337 |
-0.207 |
| 32 |
266297_at |
AT2G29570
|
ATPCNA2, A. THALIANA PROLIFERATING CELL NUCLEAR ANTIGEN 2, PCNA2, proliferating cell nuclear antigen 2 |
0.336 |
0.011 |
| 33 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.335 |
0.701 |
| 34 |
257895_at |
AT3G16950
|
LPD1, lipoamide dehydrogenase 1, ptlpd1 |
0.331 |
-0.221 |
| 35 |
250580_at |
AT5G07440
|
GDH2, glutamate dehydrogenase 2 |
0.325 |
0.649 |
| 36 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.319 |
0.551 |
| 37 |
252882_at |
AT4G39675
|
unknown |
0.317 |
0.080 |
| 38 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
0.313 |
0.043 |
| 39 |
263118_at |
AT1G03090
|
MCCA |
0.312 |
0.199 |
| 40 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
0.309 |
-0.102 |
| 41 |
263046_at |
AT2G05380
|
GRP3S, glycine-rich protein 3 short isoform |
0.307 |
0.086 |
| 42 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
0.296 |
0.445 |
| 43 |
264529_at |
AT1G30820
|
[CTP synthase family protein] |
0.295 |
-0.199 |
| 44 |
266518_at |
AT2G35170
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
0.295 |
0.052 |
| 45 |
246909_at |
AT5G25770
|
[alpha/beta-Hydrolases superfamily protein] |
0.291 |
0.248 |
| 46 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.282 |
0.601 |
| 47 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.282 |
0.097 |
| 48 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.276 |
-0.244 |
| 49 |
254420_at |
AT4G21500
|
unknown |
0.275 |
0.022 |
| 50 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.272 |
0.595 |
| 51 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.269 |
0.435 |
| 52 |
253401_at |
AT4G32870
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.268 |
0.222 |
| 53 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.713 |
-0.147 |
| 54 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.700 |
-0.383 |
| 55 |
262784_at |
AT1G10760
|
GWD, GWD1, SEX1, STARCH EXCESS 1, SOP, SOP1 |
-0.492 |
0.054 |
| 56 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.481 |
-0.137 |
| 57 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.461 |
-0.620 |
| 58 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.455 |
-0.565 |
| 59 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
-0.453 |
0.221 |
| 60 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.434 |
-0.254 |
| 61 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.432 |
-0.062 |
| 62 |
250327_at |
AT5G12050
|
unknown |
-0.425 |
-0.252 |
| 63 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
-0.414 |
0.204 |
| 64 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.399 |
-0.338 |
| 65 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.398 |
0.285 |
| 66 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.391 |
-0.146 |
| 67 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
-0.388 |
-0.210 |
| 68 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.388 |
-0.586 |
| 69 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.374 |
-0.671 |
| 70 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
-0.370 |
-0.177 |
| 71 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.364 |
-0.212 |
| 72 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
-0.357 |
-0.283 |
| 73 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.347 |
-0.138 |
| 74 |
263731_at |
AT1G59970
|
[Matrixin family protein] |
-0.342 |
0.045 |
| 75 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.341 |
-0.392 |
| 76 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.335 |
-0.398 |
| 77 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.325 |
0.651 |
| 78 |
253317_at |
AT4G33960
|
unknown |
-0.323 |
-0.001 |
| 79 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.322 |
-0.145 |
| 80 |
249325_at |
AT5G40850
|
UPM1, urophorphyrin methylase 1 |
-0.308 |
-0.038 |
| 81 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.306 |
-0.305 |
| 82 |
263883_at |
AT2G21830
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.302 |
-0.343 |
| 83 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
-0.301 |
-0.257 |
| 84 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.299 |
-0.094 |
| 85 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
-0.294 |
-0.198 |
| 86 |
258040_at |
AT3G21190
|
AtMSR1, MSR1, Mannan Synthesis Related 1 |
-0.293 |
-0.266 |
| 87 |
248085_at |
AT5G55480
|
GDPDL4, Glycerophosphodiester phosphodiesterase (GDPD) like 4, GPDL1, glycerophosphodiesterase-like 1, SVL1, SHV3-like 1 |
-0.288 |
-0.403 |
| 88 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.282 |
0.082 |
| 89 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.278 |
-0.030 |
| 90 |
251646_at |
AT3G57780
|
unknown |
-0.275 |
-0.323 |
| 91 |
261363_at |
AT1G41830
|
SKS6, SKU5 SIMILAR 6, SKS6, SKU5-similar 6 |
-0.274 |
-0.324 |
| 92 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.273 |
-0.405 |
| 93 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-0.262 |
-0.016 |
| 94 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
-0.262 |
0.418 |
| 95 |
249551_at |
AT5G38220
|
[alpha/beta-Hydrolases superfamily protein] |
-0.256 |
-0.026 |
| 96 |
262880_at |
AT1G64880
|
[Ribosomal protein S5 family protein] |
-0.252 |
-0.096 |
| 97 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.252 |
-0.496 |
| 98 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.249 |
-0.124 |
| 99 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
-0.248 |
-0.268 |
| 100 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.248 |
-0.349 |
| 101 |
251418_at |
AT3G60440
|
[Phosphoglycerate mutase family protein] |
-0.247 |
-0.132 |
| 102 |
262980_at |
AT1G75680
|
AtGH9B7, glycosyl hydrolase 9B7, GH9B7, glycosyl hydrolase 9B7 |
-0.245 |
-0.251 |
| 103 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-0.244 |
0.000 |