|
probeID |
AGICode |
Annotation |
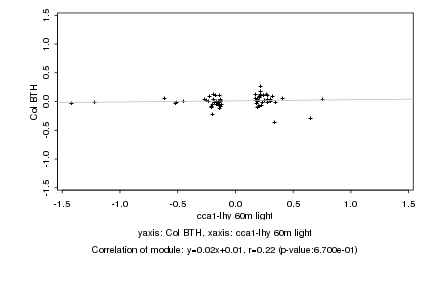
Log2 signal ratio
cca1-lhy 60m light |
Log2 signal ratio
Col BTH |
| 1 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.749 |
0.036 |
| 2 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.642 |
-0.278 |
| 3 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
0.407 |
0.066 |
| 4 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.345 |
-0.000 |
| 5 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
0.338 |
-0.351 |
| 6 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.315 |
0.097 |
| 7 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
0.302 |
0.001 |
| 8 |
257832_at |
AT3G26740
|
CCL, CCR-like |
0.301 |
0.045 |
| 9 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.275 |
0.036 |
| 10 |
265481_at |
AT2G15960
|
unknown |
0.269 |
-0.008 |
| 11 |
249811_at |
AT5G23760
|
[Copper transport protein family] |
0.269 |
0.110 |
| 12 |
251887_at |
AT3G54170
|
ATFIP37, FKBP12 interacting protein 37, FIP37, FKBP12 interacting protein 37 |
0.267 |
0.124 |
| 13 |
262958_at |
AT1G54410
|
AtHIRD11, HIRD11, dehydrin 11kDa |
0.251 |
0.030 |
| 14 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.236 |
0.119 |
| 15 |
249271_at |
AT5G41790
|
CIP1, COP1-interactive protein 1 |
0.235 |
0.120 |
| 16 |
253014_at |
AT4G37940
|
AGL21, AGAMOUS-like 21 |
0.231 |
-0.001 |
| 17 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
0.221 |
-0.067 |
| 18 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.214 |
0.269 |
| 19 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
0.212 |
0.111 |
| 20 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.212 |
0.138 |
| 21 |
252319_at |
AT3G48710
|
[DEK domain-containing chromatin associated protein] |
0.209 |
0.191 |
| 22 |
264799_at |
AT1G08550
|
AVDE1, ARABIDOPSIS VIOLAXANTHIN DE-EPOXIDASE 1, NPQ1, non-photochemical quenching 1 |
0.208 |
-0.079 |
| 23 |
247032_at |
AT5G67240
|
SDN3, small RNA degrading nuclease 3 |
0.206 |
0.072 |
| 24 |
261844_at |
AT1G15940
|
[Tudor/PWWP/MBT superfamily protein] |
0.202 |
0.094 |
| 25 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.199 |
-0.075 |
| 26 |
263023_at |
AT1G23960
|
unknown |
0.196 |
0.026 |
| 27 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.191 |
0.026 |
| 28 |
255953_at |
AT1G22070
|
TGA3, TGA1A-related gene 3 |
0.188 |
0.044 |
| 29 |
249528_at |
AT5G38720
|
unknown |
0.187 |
0.010 |
| 30 |
257791_at |
AT3G27110
|
[Peptidase family M48 family protein] |
0.186 |
0.003 |
| 31 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
0.184 |
-0.103 |
| 32 |
262170_at |
AT1G74940
|
unknown |
0.178 |
0.035 |
| 33 |
249941_at |
AT5G22270
|
unknown |
0.174 |
-0.034 |
| 34 |
245588_at |
AT4G15030
|
unknown |
0.173 |
0.126 |
| 35 |
263112_at |
AT1G03080
|
NET1D, Networked 1D |
0.166 |
0.054 |
| 36 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-1.426 |
-0.030 |
| 37 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-1.223 |
-0.009 |
| 38 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.616 |
0.061 |
| 39 |
261263_at |
AT1G26790
|
[Dof-type zinc finger DNA-binding family protein] |
-0.521 |
-0.020 |
| 40 |
256938_at |
AT3G22500
|
ATECP31, LATE EMBRYOGENESIS ABUNDANT PROTEIN ECP31 |
-0.514 |
-0.006 |
| 41 |
258575_at |
AT3G04240
|
SEC, secret agent |
-0.454 |
0.015 |
| 42 |
266757_at |
AT2G46940
|
unknown |
-0.273 |
0.038 |
| 43 |
258327_at |
AT3G22640
|
PAP85 |
-0.255 |
0.031 |
| 44 |
258576_at |
AT3G04230
|
[Ribosomal protein S5 domain 2-like superfamily protein] |
-0.238 |
0.008 |
| 45 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
-0.225 |
0.088 |
| 46 |
259268_at |
AT3G01070
|
AtENODL16, ENODL16, early nodulin-like protein 16 |
-0.208 |
-0.073 |
| 47 |
266738_at |
AT2G47010
|
unknown |
-0.208 |
-0.095 |
| 48 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.207 |
-0.221 |
| 49 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
-0.199 |
-0.044 |
| 50 |
250927_at |
AT5G03270
|
LOG6, LONELY GUY 6 |
-0.198 |
0.044 |
| 51 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.196 |
0.129 |
| 52 |
257617_at |
AT3G26550
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.185 |
-0.009 |
| 53 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
-0.177 |
0.118 |
| 54 |
245807_at |
AT1G46768
|
RAP2.1, related to AP2 1 |
-0.171 |
0.009 |
| 55 |
259782_at |
AT1G29680
|
unknown |
-0.167 |
-0.037 |
| 56 |
259189_at |
AT3G01700
|
AGP11, arabinogalactan protein 11, ATAGP11, ARABINOGALACTAN PROTEIN 11 |
-0.157 |
-0.040 |
| 57 |
252685_at |
AT3G44410
|
[pseudogene] |
-0.152 |
-0.004 |
| 58 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
-0.149 |
0.007 |
| 59 |
254152_at |
AT4G24410
|
unknown |
-0.148 |
-0.040 |
| 60 |
261997_at |
AT1G33817
|
[copia-like retrotransposon family, has a 8.1e-100 P-value blast match to gb|AAO73521.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.145 |
0.005 |
| 61 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.140 |
0.122 |
| 62 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
-0.139 |
-0.051 |
| 63 |
249450_at |
AT5G39440
|
SnRK1.3, SNF1-related protein kinase 1.3 |
-0.139 |
-0.048 |
| 64 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.139 |
-0.112 |
| 65 |
261089_at |
AT1G07570
|
APK1, APK1A |
-0.136 |
-0.071 |
| 66 |
248367_at |
AT5G52360
|
ADF10, actin depolymerizing factor 10 |
-0.132 |
0.038 |
| 67 |
250265_at |
AT5G12900
|
unknown |
-0.132 |
0.015 |
| 68 |
256281_at |
AT3G12560
|
ATTBP2, TELOMERIC DNA-BINDING PROTEIN 2, TRFL9, TRF-like 9 |
-0.130 |
0.025 |
| 69 |
263932_at |
AT2G35990
|
LOG2, LONELY GUY 2 |
-0.129 |
-0.051 |