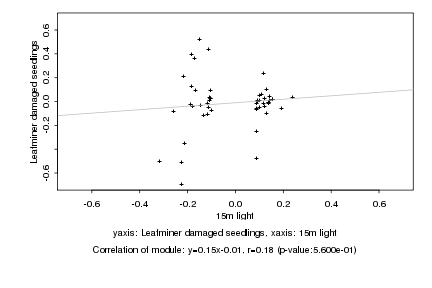
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
15m light |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.236 |
0.036 |
| 2 |
267190_at |
AT2G44170
|
ATNMT2, ARABIDOPSIS N-MYRISTOYLTRANSFERASE 2, NMT2, N-myristoyltransferase 2 |
0.190 |
-0.057 |
| 3 |
266333_at |
AT2G32410
|
AXL, AXR1-like, AXL1, AXR1- like 1 |
0.154 |
0.022 |
| 4 |
257607_at |
AT3G13880
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.152 |
0.020 |
| 5 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.142 |
0.009 |
| 6 |
267225_at |
AT2G44000
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.141 |
0.045 |
| 7 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
0.138 |
-0.005 |
| 8 |
261587_at |
AT1G01660
|
[RING/U-box superfamily protein] |
0.136 |
-0.013 |
| 9 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
0.135 |
-0.008 |
| 10 |
262268_at |
AT1G42410
|
[CACTA-like transposase family (Ptta/En/Spm), has a 1.0e-124 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.129 |
0.103 |
| 11 |
254556_at |
AT4G19220
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.127 |
-0.100 |
| 12 |
245826_at |
AT1G57850
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.121 |
0.031 |
| 13 |
254440_at |
AT4G21020
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.118 |
-0.040 |
| 14 |
265728_at |
AT2G31990
|
[Exostosin family protein] |
0.117 |
0.243 |
| 15 |
247756_at |
AT5G59100
|
[Subtilisin-like serine endopeptidase family protein] |
0.117 |
-0.011 |
| 16 |
255612_at |
AT4G01240
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.106 |
0.065 |
| 17 |
259774_at |
AT1G29520
|
[AWPM-19-like family protein] |
0.100 |
-0.049 |
| 18 |
250010_at |
AT5G18450
|
[Integrase-type DNA-binding superfamily protein] |
0.098 |
0.012 |
| 19 |
250082_at |
AT5G17200
|
[Pectin lyase-like superfamily protein] |
0.097 |
0.057 |
| 20 |
257135_at |
AT3G12900
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.092 |
0.016 |
| 21 |
254006_at |
AT4G26340
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.089 |
0.001 |
| 22 |
259531_at |
AT1G12460
|
[Leucine-rich repeat protein kinase family protein] |
0.087 |
-0.472 |
| 23 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.087 |
-0.247 |
| 24 |
263458_at |
AT2G22290
|
ATRAB-H1D, ARABIDOPSIS RAB GTPASE HOMOLOG H1D, ATRAB6, ARABIDOPSIS RAB GTPASE HOMOLOG 6, ATRABH1D, RAB GTPase homolog H1D, RAB-H1D, RAB GTPASE HOMOLOG H1D, RABH1d, RAB GTPase homolog H1D |
0.087 |
-0.014 |
| 25 |
261289_at |
AT1G37000
|
[Beta-galactosidase related protein] |
0.084 |
-0.062 |
| 26 |
254744_at |
AT4G13345
|
MEE55, maternal effect embryo arrest 55 |
0.084 |
-0.056 |
| 27 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.318 |
-0.498 |
| 28 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-0.261 |
-0.080 |
| 29 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.228 |
-0.508 |
| 30 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.228 |
-0.688 |
| 31 |
266259_at |
AT2G27830
|
unknown |
-0.219 |
0.213 |
| 32 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
-0.215 |
-0.352 |
| 33 |
249854_at |
AT5G22960
|
[alpha/beta-Hydrolases superfamily protein] |
-0.188 |
-0.022 |
| 34 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
-0.187 |
0.134 |
| 35 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
-0.184 |
0.395 |
| 36 |
256977_at |
AT3G21040
|
[copia-like retrotransposon family, has a 5.7e-20 P-value blast match to gb|AAO73527.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.181 |
-0.038 |
| 37 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.172 |
0.361 |
| 38 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.170 |
0.099 |
| 39 |
263475_at |
AT2G31945
|
unknown |
-0.152 |
0.527 |
| 40 |
249976_at |
AT5G18810
|
At-SCL28, SC35-like splicing factor 28, SCL28, SC35-like splicing factor 28 |
-0.149 |
-0.028 |
| 41 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-0.134 |
-0.115 |
| 42 |
265718_at |
AT2G03340
|
WRKY3, WRKY DNA-binding protein 3 |
-0.118 |
-0.101 |
| 43 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
-0.117 |
-0.011 |
| 44 |
247800_at |
AT5G58570
|
unknown |
-0.116 |
-0.044 |
| 45 |
264774_at |
AT1G22890
|
unknown |
-0.115 |
0.444 |
| 46 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
-0.112 |
0.034 |
| 47 |
267255_at |
AT2G22950
|
ACA7, auto-regulated Ca2+-ATPase 7 |
-0.110 |
0.012 |
| 48 |
246203_at |
AT4G36610
|
[alpha/beta-Hydrolases superfamily protein] |
-0.107 |
0.033 |
| 49 |
257469_at |
AT1G49290
|
unknown |
-0.107 |
0.029 |
| 50 |
260256_at |
AT1G74350
|
[Intron maturase, type II family protein] |
-0.105 |
0.095 |
| 51 |
260366_at |
AT1G70460
|
AtPERK13, proline-rich extensin-like receptor kinase 13, PERK13, proline-rich extensin-like receptor kinase 13, RHS10, root hair specific 10 |
-0.104 |
-0.072 |