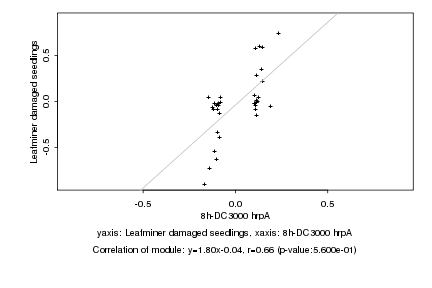
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
8h-DC3000 hrpA |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
0.233 |
0.744 |
| 2 |
258427_at |
AT3G16600
|
[SNF2 domain-containing protein / helicase domain-containing protein / zinc finger protein-related] |
0.186 |
-0.050 |
| 3 |
254996_at |
AT4G10390
|
[Protein kinase superfamily protein] |
0.145 |
0.592 |
| 4 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
0.144 |
0.220 |
| 5 |
261366_at |
AT1G53100
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.136 |
0.351 |
| 6 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.129 |
0.603 |
| 7 |
251875_at |
AT3G54270
|
[sucrose-6F-phosphate phosphohydrolase family protein] |
0.123 |
0.052 |
| 8 |
262446_at |
AT1G49310
|
unknown |
0.115 |
0.001 |
| 9 |
254881_at |
AT4G11700
|
unknown |
0.114 |
0.013 |
| 10 |
249449_at |
AT5G39430
|
unknown |
0.113 |
-0.007 |
| 11 |
251501_at |
AT3G59120
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.110 |
-0.009 |
| 12 |
260361_at |
AT1G69360
|
unknown |
0.109 |
0.286 |
| 13 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.109 |
-0.142 |
| 14 |
262061_at |
AT1G80110
|
ATPP2-B11, phloem protein 2-B11, PP2-B11, phloem protein 2-B11 |
0.108 |
0.583 |
| 15 |
247970_at |
AT5G56720
|
c-NAD-MDH3, cytosolic-NAD-dependent malate dehydrogenase 3 |
0.105 |
-0.076 |
| 16 |
265859_at |
AT2G01700
|
unknown |
0.105 |
-0.041 |
| 17 |
254339_at |
AT4G22100
|
BGLU3, beta glucosidase 2 |
0.100 |
-0.021 |
| 18 |
255204_at |
AT4G07530
|
unknown |
0.098 |
-0.018 |
| 19 |
246183_at |
AT5G20940
|
[Glycosyl hydrolase family protein] |
0.098 |
0.067 |
| 20 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.171 |
-0.892 |
| 21 |
253067_at |
AT4G37780
|
ATMYB87, MYB87, myb domain protein 87 |
-0.146 |
0.048 |
| 22 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.143 |
-0.720 |
| 23 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.129 |
-0.057 |
| 24 |
264357_at |
AT1G03360
|
ATRRP4, ribosomal RNA processing 4, RRP4, ribosomal RNA processing 4 |
-0.121 |
-0.085 |
| 25 |
255149_at |
AT4G08150
|
BP, BREVIPEDICELLUS, BP1, BREVIPEDICELLUS 1, KNAT1, KNOTTED-like from Arabidopsis thaliana |
-0.115 |
-0.019 |
| 26 |
259760_at |
AT1G77580
|
unknown |
-0.114 |
-0.535 |
| 27 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.108 |
-0.621 |
| 28 |
251079_at |
AT5G02000
|
unknown |
-0.106 |
-0.033 |
| 29 |
249546_at |
AT5G38150
|
PMI15, plastid movement impaired 15 |
-0.102 |
-0.328 |
| 30 |
257931_at |
AT3G17030
|
[Nucleic acid-binding proteins superfamily] |
-0.099 |
-0.024 |
| 31 |
253802_at |
AT4G28180
|
unknown |
-0.098 |
-0.079 |
| 32 |
249186_at |
AT5G43040
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.097 |
-0.037 |
| 33 |
267371_at |
AT2G44510
|
[CDK inhibitor P21 binding protein] |
-0.092 |
-0.020 |
| 34 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
-0.091 |
-0.388 |
| 35 |
250674_at |
AT5G07130
|
LAC13, laccase 13 |
-0.090 |
-0.126 |
| 36 |
250022_at |
AT5G18210
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.085 |
0.051 |
| 37 |
246834_at |
AT5G26630
|
[MADS-box transcription factor family protein] |
-0.083 |
-0.003 |