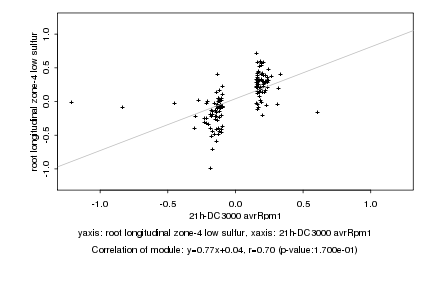
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
21h-DC3000 avrRpm1 |
Log2 signal ratio
root longitudinal zone-4 low sulfur |
| 1 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
0.601 |
-0.158 |
| 2 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
0.327 |
0.414 |
| 3 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.311 |
0.199 |
| 4 |
257636_at |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
0.304 |
-0.030 |
| 5 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
0.260 |
0.376 |
| 6 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
0.244 |
0.483 |
| 7 |
258505_at |
AT3G06530
|
[ARM repeat superfamily protein] |
0.241 |
0.325 |
| 8 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
0.236 |
0.326 |
| 9 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
0.236 |
0.291 |
| 10 |
256797_at |
AT3G18600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.231 |
0.210 |
| 11 |
258009_at |
AT3G19440
|
[Pseudouridine synthase family protein] |
0.230 |
0.363 |
| 12 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.225 |
-0.059 |
| 13 |
256288_at |
AT3G12270
|
ATPRMT3, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 3, PRMT3, protein arginine methyltransferase 3 |
0.225 |
0.310 |
| 14 |
249354_at |
AT5G40480
|
EMB3012, embryo defective 3012 |
0.217 |
0.169 |
| 15 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
0.217 |
0.311 |
| 16 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
0.215 |
0.400 |
| 17 |
251611_at |
AT3G57940
|
unknown |
0.210 |
0.272 |
| 18 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.208 |
0.135 |
| 19 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
0.203 |
0.232 |
| 20 |
264357_at |
AT1G03360
|
ATRRP4, ribosomal RNA processing 4, RRP4, ribosomal RNA processing 4 |
0.202 |
0.591 |
| 21 |
267349_at |
AT2G40010
|
[Ribosomal protein L10 family protein] |
0.201 |
0.299 |
| 22 |
263864_at |
AT2G04530
|
CPZ, TRZ2, TRNASE Z 2 |
0.201 |
0.254 |
| 23 |
253492_at |
AT4G31810
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.200 |
0.389 |
| 24 |
265991_at |
AT2G24120
|
PDE319, PIGMENT DEFECTIVE 319, SCA3, SCABRA 3 |
0.199 |
0.134 |
| 25 |
257667_at |
AT3G20440
|
BE1, BRANCHING ENZYME 1, EMB2729, EMBRYO DEFECTIVE 2729 |
0.198 |
0.141 |
| 26 |
251354_at |
AT3G61090
|
[Putative endonuclease or glycosyl hydrolase] |
0.195 |
-0.195 |
| 27 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.194 |
0.422 |
| 28 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
0.193 |
0.307 |
| 29 |
254043_at |
AT4G25990
|
CIL |
0.191 |
-0.006 |
| 30 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
0.190 |
0.405 |
| 31 |
253598_at |
AT4G30800
|
[Nucleic acid-binding, OB-fold-like protein] |
0.190 |
0.570 |
| 32 |
265326_at |
AT2G18220
|
[Noc2p family] |
0.189 |
0.341 |
| 33 |
251476_at |
AT3G59670
|
unknown |
0.188 |
0.539 |
| 34 |
246303_at |
AT3G51870
|
[Mitochondrial substrate carrier family protein] |
0.186 |
0.324 |
| 35 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
0.183 |
0.192 |
| 36 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
0.183 |
0.215 |
| 37 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
0.182 |
0.595 |
| 38 |
261775_at |
AT1G76280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.178 |
0.017 |
| 39 |
251256_at |
AT3G62300
|
unknown |
0.176 |
0.081 |
| 40 |
247739_at |
AT5G59240
|
[Ribosomal protein S8e family protein] |
0.174 |
0.524 |
| 41 |
256077_at |
AT1G18090
|
[5'-3' exonuclease family protein] |
0.174 |
0.150 |
| 42 |
260316_at |
AT1G63810
|
unknown |
0.172 |
0.324 |
| 43 |
255225_at |
AT4G05410
|
YAO, YAOZHE |
0.171 |
0.182 |
| 44 |
253057_at |
AT4G37670
|
NAGS2, N-acetyl-l-glutamate synthase 2 |
0.171 |
0.141 |
| 45 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
0.170 |
-0.081 |
| 46 |
254076_at |
AT4G25340
|
ATFKBP53, FKBP53, FK506 BINDING PROTEIN 53 |
0.168 |
0.312 |
| 47 |
265730_at |
AT2G32220
|
[Ribosomal L27e protein family] |
0.168 |
0.262 |
| 48 |
247032_at |
AT5G67240
|
SDN3, small RNA degrading nuclease 3 |
0.167 |
0.438 |
| 49 |
261296_at |
AT1G48460
|
unknown |
0.166 |
0.316 |
| 50 |
252508_at |
AT3G46210
|
[Ribosomal protein S5 domain 2-like superfamily protein] |
0.165 |
0.456 |
| 51 |
245502_at |
AT4G15640
|
unknown |
0.163 |
0.319 |
| 52 |
256661_at |
AT3G11964
|
RRP5, Ribosomal RNA Processing 5 |
0.162 |
0.237 |
| 53 |
265139_at |
AT1G51310
|
[transferases] |
0.162 |
0.162 |
| 54 |
254173_at |
AT4G24580
|
REN1, ROP1 ENHANCER 1 |
0.162 |
-0.113 |
| 55 |
265274_at |
AT2G28450
|
[zinc finger (CCCH-type) family protein] |
0.161 |
0.417 |
| 56 |
255756_at |
AT1G19940
|
AtGH9B5, glycosyl hydrolase 9B5, GH9B5, glycosyl hydrolase 9B5 |
0.160 |
-0.037 |
| 57 |
248984_at |
AT5G45140
|
NRPC2, nuclear RNA polymerase C2 |
0.160 |
0.132 |
| 58 |
264818_at |
AT1G03530
|
ATNAF1, NAF1, nuclear assembly factor 1 |
0.159 |
0.393 |
| 59 |
245909_at |
AT5G09380
|
[RNA polymerase III RPC4] |
0.159 |
0.254 |
| 60 |
248357_at |
AT5G52380
|
[VASCULAR-RELATED NAC-DOMAIN 6] |
0.158 |
0.289 |
| 61 |
246457_at |
AT5G16750
|
TOZ, TORMOZEMBRYO DEFECTIVE |
0.158 |
0.586 |
| 62 |
261301_at |
AT1G48570
|
[zinc finger (Ran-binding) family protein] |
0.158 |
0.222 |
| 63 |
260157_at |
AT1G52930
|
[Ribosomal RNA processing Brix domain protein] |
0.158 |
0.358 |
| 64 |
260235_at |
AT1G74560
|
NRP1, NAP1-related protein 1 |
0.157 |
0.160 |
| 65 |
252410_at |
AT3G47450
|
ATNOA1, ATNOS1, NOA1, NO ASSOCIATED 1, NOS1, NITRIC OXIDE SYNTHASE 1, RIF1, RESISTANT TO INHIBITION WITH FOSMIDOMYCIN 1 |
0.155 |
0.216 |
| 66 |
267044_at |
AT2G34357
|
[ARM repeat superfamily protein] |
0.154 |
0.293 |
| 67 |
258111_at |
AT3G14630
|
CYP72A9, cytochrome P450, family 72, subfamily A, polypeptide 9 |
0.152 |
-0.029 |
| 68 |
263843_at |
AT2G37020
|
[Translin family protein] |
0.152 |
0.230 |
| 69 |
255501_at |
AT4G02400
|
[U3 ribonucleoprotein (Utp) family protein] |
0.151 |
0.329 |
| 70 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
0.150 |
0.717 |
| 71 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
-1.217 |
-0.002 |
| 72 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.839 |
-0.088 |
| 73 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.451 |
-0.019 |
| 74 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
-0.307 |
-0.397 |
| 75 |
254996_at |
AT4G10390
|
[Protein kinase superfamily protein] |
-0.300 |
-0.214 |
| 76 |
247730_at |
AT5G59580
|
UGT76E1, UDP-glucosyl transferase 76E1 |
-0.280 |
0.017 |
| 77 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
-0.236 |
-0.248 |
| 78 |
247882_at |
AT5G57785
|
unknown |
-0.231 |
-0.310 |
| 79 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.219 |
-0.326 |
| 80 |
255912_at |
AT1G66960
|
LUP5 |
-0.218 |
-0.029 |
| 81 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.217 |
-0.244 |
| 82 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.211 |
0.008 |
| 83 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
-0.200 |
-0.339 |
| 84 |
246197_at |
AT4G37010
|
CEN2, centrin 2 |
-0.187 |
-0.983 |
| 85 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
-0.185 |
-0.397 |
| 86 |
263972_at |
AT2G42760
|
unknown |
-0.185 |
-0.186 |
| 87 |
252084_at |
AT3G51970
|
ASAT1, acyl-CoA sterol acyl transferase 1, ATASAT1, ATSAT1, ARABIDOPSIS THALIANA STEROL O-ACYLTRANSFERASE 1 |
-0.183 |
-0.119 |
| 88 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
-0.182 |
-0.209 |
| 89 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-0.181 |
-0.509 |
| 90 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
-0.174 |
-0.441 |
| 91 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
-0.174 |
-0.705 |
| 92 |
249910_at |
AT5G22630
|
ADT5, arogenate dehydratase 5 |
-0.170 |
-0.179 |
| 93 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
-0.170 |
-0.146 |
| 94 |
262811_at |
AT1G11700
|
unknown |
-0.156 |
-0.482 |
| 95 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.156 |
-0.019 |
| 96 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
-0.154 |
-0.260 |
| 97 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
-0.153 |
-0.133 |
| 98 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.150 |
-0.216 |
| 99 |
247304_at |
AT5G63850
|
AAP4, amino acid permease 4 |
-0.146 |
-0.584 |
| 100 |
255036_at |
AT4G09560
|
[Protease-associated (PA) RING/U-box zinc finger family protein] |
-0.144 |
-0.407 |
| 101 |
264993_at |
AT1G67290
|
GLOX1, glyoxal oxidase 1 |
-0.142 |
-0.158 |
| 102 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
-0.141 |
-0.209 |
| 103 |
260887_at |
AT1G29160
|
[Dof-type zinc finger DNA-binding family protein] |
-0.141 |
-0.073 |
| 104 |
248133_at |
AT5G54850
|
unknown |
-0.140 |
0.139 |
| 105 |
256947_at |
AT3G19050
|
POK2, phragmoplast orienting kinesin 2 |
-0.139 |
-0.033 |
| 106 |
251080_at |
AT5G02010
|
ATROPGEF7, ROPGEF7, ROP (rho of plants) guanine nucleotide exchange factor 7 |
-0.135 |
0.410 |
| 107 |
259708_at |
AT1G77420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.135 |
-0.095 |
| 108 |
258752_at |
AT3G09520
|
ATEXO70H4, exocyst subunit exo70 family protein H4, EXO70H4, exocyst subunit exo70 family protein H4 |
-0.134 |
-0.274 |
| 109 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
-0.132 |
0.055 |
| 110 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.131 |
-0.480 |
| 111 |
255961_at |
AT1G22340
|
AtUGT85A7, UDP-glucosyl transferase 85A7, UGT85A7, UDP-glucosyl transferase 85A7 |
-0.130 |
0.014 |
| 112 |
246370_at |
AT1G51920
|
unknown |
-0.126 |
-0.395 |
| 113 |
247424_at |
AT5G62850
|
AtSWEET5, AtVEX1, VEGETATIVE CELL EXPRESSED1, SWEET5 |
-0.126 |
0.045 |
| 114 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
-0.125 |
-0.225 |
| 115 |
251916_at |
AT3G53960
|
[Major facilitator superfamily protein] |
-0.124 |
0.031 |
| 116 |
256572_at |
AT3G30740
|
[pseudogene] |
-0.124 |
0.012 |
| 117 |
251826_at |
AT3G55110
|
ABCG18, ATP-binding cassette G18 |
-0.124 |
-0.086 |
| 118 |
249889_at |
AT5G22540
|
unknown |
-0.124 |
-0.084 |
| 119 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
-0.124 |
-0.441 |
| 120 |
260897_at |
AT1G29330
|
AERD2, ARABIDOPSIS ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ATERD2, ARABIDOPSIS THALIANA ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ERD2, ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2 |
-0.122 |
0.169 |
| 121 |
265479_at |
AT2G15760
|
unknown |
-0.121 |
-0.064 |
| 122 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.120 |
-0.060 |
| 123 |
260581_at |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
-0.119 |
-0.142 |
| 124 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
-0.118 |
0.026 |
| 125 |
256433_at |
AT3G10985
|
ATWI-12, ARABIDOPSIS THALIANA WOUND-INDUCED PROTEIN 12, SAG20, senescence associated gene 20, WI12, WOUND-INDUCED PROTEIN 12 |
-0.115 |
-0.058 |
| 126 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
-0.111 |
-0.039 |
| 127 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.111 |
-0.100 |
| 128 |
258201_at |
AT3G13910
|
unknown |
-0.107 |
-0.453 |
| 129 |
254416_at |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
-0.107 |
0.057 |
| 130 |
265057_at |
AT1G52140
|
unknown |
-0.107 |
-0.415 |
| 131 |
260259_at |
AT1G74300
|
[alpha/beta-Hydrolases superfamily protein] |
-0.106 |
0.029 |
| 132 |
250948_at |
AT5G03490
|
[UDP-Glycosyltransferase superfamily protein] |
-0.106 |
-0.196 |
| 133 |
249728_at |
AT5G24390
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
-0.103 |
0.232 |
| 134 |
246046_at |
AT5G28860
|
unknown |
-0.103 |
-0.085 |
| 135 |
267393_at |
AT2G44500
|
[O-fucosyltransferase family protein] |
-0.100 |
-0.074 |
| 136 |
253401_at |
AT4G32870
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.099 |
-0.063 |
| 137 |
261754_at |
AT1G76130
|
AMY2, alpha-amylase-like 2, ATAMY2, ARABIDOPSIS THALIANA ALPHA-AMYLASE-LIKE 2 |
-0.098 |
-0.072 |
| 138 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
-0.098 |
-0.364 |
| 139 |
256292_at |
AT1G69430
|
unknown |
-0.098 |
0.112 |