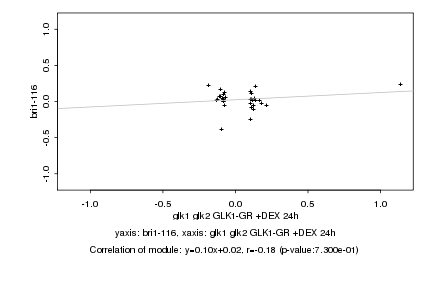
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
glk1 glk2 GLK1-GR +DEX 24h |
Log2 signal ratio
bri1-116 |
| 1 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
1.135 |
0.241 |
| 2 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
0.212 |
-0.043 |
| 3 |
260768_at |
AT1G49245
|
[Prefoldin chaperone subunit family protein] |
0.177 |
-0.017 |
| 4 |
266465_at |
AT2G47750
|
GH3.9, putative indole-3-acetic acid-amido synthetase GH3.9 |
0.160 |
0.021 |
| 5 |
247737_at |
AT5G59200
|
OTP80, ORGANELLE TRANSCRIPT PROCESSING 80 |
0.136 |
0.210 |
| 6 |
259857_at |
AT1G68430
|
unknown |
0.132 |
0.026 |
| 7 |
245636_at |
AT1G25240
|
[ENTH/VHS/GAT family protein] |
0.131 |
0.042 |
| 8 |
267538_at |
AT2G41870
|
[Remorin family protein] |
0.124 |
-0.106 |
| 9 |
245161_at |
AT2G33070
|
ATNSP2, NITRILE-SPECIFIER PROTEIN 2, NSP2, nitrile specifier protein 2 |
0.120 |
-0.043 |
| 10 |
249507_at |
AT5G38370
|
[FUNCTIONS IN: zinc ion binding] |
0.116 |
0.027 |
| 11 |
267522_at |
AT2G30430
|
unknown |
0.107 |
0.030 |
| 12 |
252395_at |
AT3G47950
|
AHA4, H(+)-ATPase 4, HA4, H(+)-ATPase 4 |
0.105 |
-0.073 |
| 13 |
266592_at |
AT2G46210
|
AtSLD2, SLD2, sphingoid LCB desaturase 2 |
0.104 |
0.122 |
| 14 |
256097_at |
AT1G13670
|
unknown |
0.101 |
-0.024 |
| 15 |
245546_at |
AT4G15290
|
ATCSLB05, ATCSLB5, CELLULOSE SYNTHASE LIKE 5, CSLB05 |
0.101 |
-0.240 |
| 16 |
255839_at |
AT2G33500
|
BBX12, B-box domain protein 12 |
0.100 |
0.152 |
| 17 |
266431_at |
AT2G07230
|
[Mutator-like transposase family, has a 8.1e-59 P-value blast match to Q9SHN7 /450-633 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
0.098 |
0.041 |
| 18 |
256484_at |
AT1G31430
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.188 |
0.225 |
| 19 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.135 |
0.015 |
| 20 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
-0.128 |
0.033 |
| 21 |
252771_at |
AT3G42880
|
PRK3, pollen receptor like kinase 3 |
-0.115 |
0.071 |
| 22 |
247110_at |
AT5G65830
|
ATRLP57, receptor like protein 57, RLP57, receptor like protein 57 |
-0.109 |
0.075 |
| 23 |
245208_at |
AT5G12330
|
LRP1, LATERAL ROOT PRIMORDIUM 1 |
-0.103 |
0.177 |
| 24 |
263178_at |
AT1G05550
|
unknown |
-0.100 |
-0.381 |
| 25 |
265189_at |
AT1G23840
|
unknown |
-0.090 |
0.050 |
| 26 |
265881_at |
AT2G42480
|
[TRAF-like family protein] |
-0.089 |
0.031 |
| 27 |
251188_at |
AT3G62730
|
unknown |
-0.088 |
0.035 |
| 28 |
257449_at |
AT2G31420
|
[FUNCTIONS IN: DNA binding] |
-0.087 |
0.004 |
| 29 |
265621_at |
AT2G27300
|
ANAC040, Arabidopsis NAC domain containing protein 40, NTL8, NTM1-like 8 |
-0.085 |
0.099 |
| 30 |
247141_at |
AT5G65560
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.083 |
0.008 |
| 31 |
245640_at |
AT1G25330
|
CES, CESTA, HAF, HALF FILLED |
-0.081 |
0.132 |
| 32 |
261599_at |
AT1G49700
|
unknown |
-0.076 |
-0.046 |
| 33 |
245634_at |
AT1G25270
|
UMAMIT24, Usually multiple acids move in and out Transporters 24 |
-0.075 |
0.067 |