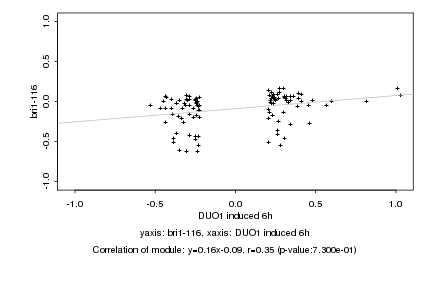
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
DUO1 induced 6h |
Log2 signal ratio
bri1-116 |
| 1 |
255815_at |
AT1G19890
|
ATMGH3, MALE-GAMETE-SPECIFIC HISTONE H3, MGH3, male-gamete-specific histone H3 |
1.026 |
0.085 |
| 2 |
252440_at |
AT3G47440
|
TIP5;1, tonoplast intrinsic protein 5;1 |
1.004 |
0.164 |
| 3 |
249468_at |
AT5G39650
|
unknown |
0.811 |
0.006 |
| 4 |
261670_at |
AT1G18520
|
TET11, tetraspanin11 |
0.594 |
0.006 |
| 5 |
250535_at |
AT5G08480
|
[VQ motif-containing protein] |
0.562 |
-0.049 |
| 6 |
248011_at |
AT5G56300
|
GAMT2, gibberellic acid methyltransferase 2 |
0.480 |
0.023 |
| 7 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.458 |
-0.263 |
| 8 |
258840_at |
AT3G04620
|
DAN1, D NUCLDUO1-ACTIVATEEIC ACID BINDING PROTEIN 1 |
0.452 |
-0.044 |
| 9 |
247758_at |
AT5G59120
|
ATSBT4.13, subtilase 4.13, SBT4.13, subtilase 4.13 |
0.408 |
0.098 |
| 10 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
0.407 |
0.012 |
| 11 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.392 |
0.041 |
| 12 |
261396_at |
AT1G79800
|
AtENODL7, ENODL7, early nodulin-like protein 7 |
0.389 |
0.103 |
| 13 |
248553_at |
AT5G50170
|
[C2 calcium/lipid-binding and GRAM domain containing protein] |
0.384 |
-0.057 |
| 14 |
249617_at |
AT5G37450
|
[Leucine-rich repeat protein kinase family protein] |
0.360 |
0.072 |
| 15 |
256684_at |
AT3G32040
|
[Terpenoid synthases superfamily protein] |
0.343 |
0.069 |
| 16 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
0.341 |
0.016 |
| 17 |
260648_at |
AT1G08050
|
[Zinc finger (C3HC4-type RING finger) family protein] |
0.341 |
-0.287 |
| 18 |
248275_at |
AT5G53520
|
ATOPT8, oligopeptide transporter 8, OPT8, oligopeptide transporter 8 |
0.326 |
-0.007 |
| 19 |
246459_at |
AT5G16900
|
[Leucine-rich repeat protein kinase family protein] |
0.316 |
0.022 |
| 20 |
248443_at |
AT5G51310
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.312 |
0.075 |
| 21 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
0.311 |
0.040 |
| 22 |
246386_at |
AT1G77410
|
BGAL16, beta-galactosidase 16 |
0.311 |
0.049 |
| 23 |
253774_at |
AT4G28530
|
anac074, NAC domain containing protein 74, NAC074, NAC domain containing protein 74 |
0.302 |
-0.461 |
| 24 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
0.301 |
0.063 |
| 25 |
250051_at |
AT5G17800
|
AtMYB56, myb domain protein 56, MYB56, myb domain protein 56 |
0.300 |
0.060 |
| 26 |
261027_at |
AT1G01340
|
ACBK1, ATCNGC10, cyclic nucleotide gated channel 10, CNGC10, cyclic nucleotide gated channel 10 |
0.297 |
-0.129 |
| 27 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
0.295 |
0.167 |
| 28 |
262105_at |
AT1G02810
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.275 |
-0.549 |
| 29 |
262606_at |
AT1G15190
|
[Fasciclin-like arabinogalactan family protein] |
0.273 |
0.115 |
| 30 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.269 |
0.167 |
| 31 |
254689_at |
AT4G13710
|
[Pectin lyase-like superfamily protein] |
0.268 |
-0.248 |
| 32 |
252986_at |
AT4G38380
|
[MATE efflux family protein] |
0.265 |
0.044 |
| 33 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.262 |
-0.356 |
| 34 |
252248_at |
AT3G49750
|
AtRLP44, receptor like protein 44, RLP44, receptor like protein 44 |
0.259 |
-0.411 |
| 35 |
262115_at |
AT1G02813
|
unknown |
0.257 |
0.092 |
| 36 |
248167_at |
AT5G54530
|
unknown |
0.249 |
0.018 |
| 37 |
260145_at |
AT1G52920
|
GCR2, G-PROTEIN COUPLED RECEPTOR 2, GPCR, G protein coupled receptor |
0.248 |
0.035 |
| 38 |
251130_at |
AT5G01180
|
AtNPF8.2, ATPTR5, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 5, NPF8.2, NRT1/ PTR family 8.2, PTR5, peptide transporter 5 |
0.239 |
0.054 |
| 39 |
261198_at |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
0.236 |
0.091 |
| 40 |
258172_at |
AT3G21620
|
[ERD (early-responsive to dehydration stress) family protein] |
0.235 |
-0.015 |
| 41 |
251761_at |
AT3G55700
|
[UDP-Glycosyltransferase superfamily protein] |
0.234 |
0.040 |
| 42 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
0.228 |
-0.165 |
| 43 |
266689_at |
AT2G19930
|
[RNA-dependent RNA polymerase family protein] |
0.228 |
0.067 |
| 44 |
258863_at |
AT3G03300
|
ATDCL2, DICER-LIKE 2, DCL2, dicer-like 2 |
0.224 |
0.116 |
| 45 |
262664_at |
AT1G13970
|
unknown |
0.223 |
0.042 |
| 46 |
262358_at |
AT1G73050
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.219 |
-0.014 |
| 47 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
0.217 |
-0.012 |
| 48 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
0.215 |
0.024 |
| 49 |
263417_at |
AT2G17180
|
DAZ1, DUO1-ACTIVATED ZINC FINGER 1 |
0.209 |
-0.126 |
| 50 |
267426_at |
AT2G34820
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.209 |
0.085 |
| 51 |
265031_at |
AT1G61590
|
[Protein kinase superfamily protein] |
0.205 |
-0.093 |
| 52 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
0.204 |
-0.507 |
| 53 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.204 |
0.146 |
| 54 |
258745_at |
AT3G05920
|
[Heavy metal transport/detoxification superfamily protein ] |
0.201 |
-0.208 |
| 55 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.533 |
-0.046 |
| 56 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.472 |
-0.081 |
| 57 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.451 |
0.012 |
| 58 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
-0.440 |
-0.257 |
| 59 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.439 |
0.069 |
| 60 |
255695_at |
AT4G00080
|
UNE11, unfertilized embryo sac 11 |
-0.437 |
-0.078 |
| 61 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
-0.430 |
0.057 |
| 62 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
-0.404 |
-0.082 |
| 63 |
245172_at |
AT2G47540
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.402 |
0.032 |
| 64 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
-0.398 |
-0.151 |
| 65 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.389 |
-0.503 |
| 66 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
-0.388 |
-0.455 |
| 67 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.373 |
-0.013 |
| 68 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.368 |
-0.018 |
| 69 |
252511_at |
AT3G46280
|
[protein kinase-related] |
-0.368 |
-0.397 |
| 70 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.355 |
-0.177 |
| 71 |
262045_at |
AT1G80240
|
DGR1, DUF642 L-GalL responsive gene 1 |
-0.351 |
-0.602 |
| 72 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.349 |
0.025 |
| 73 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
-0.337 |
-0.209 |
| 74 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
-0.336 |
-0.087 |
| 75 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
-0.329 |
-0.261 |
| 76 |
250469_at |
AT5G10130
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.320 |
-0.019 |
| 77 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.312 |
-0.038 |
| 78 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.311 |
-0.614 |
| 79 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
-0.310 |
0.082 |
| 80 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.306 |
0.028 |
| 81 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.301 |
0.021 |
| 82 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.292 |
-0.162 |
| 83 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
-0.289 |
0.028 |
| 84 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
-0.289 |
-0.414 |
| 85 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
-0.288 |
0.065 |
| 86 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
-0.287 |
-0.049 |
| 87 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
-0.266 |
-0.198 |
| 88 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.262 |
-0.086 |
| 89 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
-0.255 |
-0.469 |
| 90 |
254673_at |
AT4G18430
|
AtRABA1e, RAB GTPase homolog A1E, RABA1e, RAB GTPase homolog A1E |
-0.251 |
-0.431 |
| 91 |
266784_at |
AT2G28960
|
[Leucine-rich repeat protein kinase family protein] |
-0.251 |
0.029 |
| 92 |
246366_at |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
-0.250 |
0.021 |
| 93 |
267222_at |
AT2G43880
|
[Pectin lyase-like superfamily protein] |
-0.246 |
0.049 |
| 94 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.245 |
-0.021 |
| 95 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.244 |
-0.168 |
| 96 |
250778_at |
AT5G05500
|
MOP10 |
-0.243 |
-0.008 |
| 97 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.241 |
-0.618 |
| 98 |
247645_at |
AT5G60530
|
[late embryogenesis abundant protein-related / LEA protein-related] |
-0.241 |
0.004 |
| 99 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.239 |
-0.039 |
| 100 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
-0.236 |
-0.104 |
| 101 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
-0.234 |
-0.433 |
| 102 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.232 |
-0.538 |
| 103 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.232 |
-0.061 |
| 104 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
-0.229 |
0.061 |
| 105 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.227 |
-0.043 |
| 106 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.226 |
-0.104 |
| 107 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-0.226 |
-0.192 |