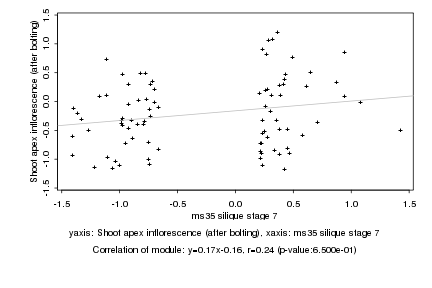
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ms35 silique stage 7 |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
261264_at |
AT1G26710
|
unknown |
1.422 |
-0.501 |
| 2 |
262421_at |
AT1G50290
|
unknown |
1.073 |
-0.003 |
| 3 |
249656_at |
AT5G37130
|
[Protein prenylyltransferase superfamily protein] |
0.935 |
0.098 |
| 4 |
246731_at |
AT5G27630
|
ACBP5, acyl-CoA binding protein 5, AtACBP5 |
0.934 |
0.864 |
| 5 |
263023_at |
AT1G23960
|
unknown |
0.867 |
0.331 |
| 6 |
250135_at |
AT5G15360
|
unknown |
0.707 |
-0.350 |
| 7 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.640 |
0.509 |
| 8 |
262458_at |
AT1G11280
|
[S-locus lectin protein kinase family protein] |
0.610 |
0.265 |
| 9 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.574 |
-0.577 |
| 10 |
246870_at |
AT5G26030
|
ATFC-I, FC-I, FC1, ferrochelatase 1 |
0.486 |
0.769 |
| 11 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
0.460 |
-0.892 |
| 12 |
246542_at |
AT5G15020
|
SNL2, SIN3-like 2 |
0.449 |
-0.810 |
| 13 |
259100_at |
AT3G04880
|
DRT102, DNA-DAMAGE-REPAIR/TOLERATION 2 |
0.442 |
-0.472 |
| 14 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
0.428 |
0.481 |
| 15 |
252347_at |
AT3G48130
|
RSU1 |
0.423 |
-1.165 |
| 16 |
264835_at |
AT1G03550
|
AtSCAMP4, SCAMP4, Secretory carrier membrane protein 4 |
0.419 |
0.390 |
| 17 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.411 |
0.310 |
| 18 |
250530_at |
AT5G08630
|
[DDT domain-containing protein] |
0.384 |
0.115 |
| 19 |
246080_at |
AT5G20460
|
unknown |
0.380 |
-0.907 |
| 20 |
249016_at |
AT5G44750
|
ATREV1, ARABIDOPSIS THALIANA HOMOLOG OF REVERSIONLESS 1, REV1 |
0.375 |
-0.479 |
| 21 |
252676_at |
AT3G44280
|
unknown |
0.375 |
0.287 |
| 22 |
266587_at |
AT2G14880
|
[SWIB/MDM2 domain superfamily protein] |
0.360 |
1.202 |
| 23 |
247754_at |
AT5G59080
|
unknown |
0.354 |
-0.322 |
| 24 |
249386_at |
AT5G40060
|
[Disease resistance protein (NBS-LRR class) family] |
0.336 |
-0.834 |
| 25 |
261248_at |
AT1G20030
|
[Pathogenesis-related thaumatin superfamily protein] |
0.318 |
1.084 |
| 26 |
250699_at |
AT5G06820
|
SRF2, STRUBBELIG-receptor family 2 |
0.304 |
0.107 |
| 27 |
263258_at |
AT1G10540
|
ATNAT8, NAT8, nucleobase-ascorbate transporter 8 |
0.298 |
-0.161 |
| 28 |
261681_at |
AT1G47340
|
[F-box and associated interaction domains-containing protein] |
0.280 |
1.059 |
| 29 |
257136_at |
AT3G12890
|
ASML2, activator of spomin::LUC2 |
0.272 |
-0.616 |
| 30 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
0.269 |
0.221 |
| 31 |
250483_at |
AT5G10300
|
AtHNL, ATMES5, ARABIDOPSIS THALIANA METHYL ESTERASE 5, HNL, HYDROXYNITRILE LYASE, MES5, methyl esterase 5 |
0.266 |
0.823 |
| 32 |
252351_at |
AT3G48210
|
unknown |
0.258 |
-0.069 |
| 33 |
247456_at |
AT5G62160
|
AtZIP12, zinc transporter 12 precursor, ZIP12, zinc transporter 12 precursor |
0.257 |
0.203 |
| 34 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.249 |
-0.516 |
| 35 |
265771_at |
AT2G48030
|
[DNAse I-like superfamily protein] |
0.233 |
-0.317 |
| 36 |
249493_at |
AT5G39080
|
[HXXXD-type acyl-transferase family protein] |
0.232 |
-1.108 |
| 37 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.230 |
-0.543 |
| 38 |
252315_at |
AT3G48690
|
ATCXE12, ARABIDOPSIS THALIANA CARBOXYESTERASE 12, CXE12 |
0.227 |
0.919 |
| 39 |
247967_at |
AT5G56620
|
anac099, NAC domain containing protein 99, NAC099, NAC domain containing protein 99 |
0.222 |
-0.718 |
| 40 |
262302_at |
AT1G70910
|
DEP, DESPIERTO |
0.221 |
-0.892 |
| 41 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
0.212 |
-0.859 |
| 42 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.209 |
-0.988 |
| 43 |
266125_at |
AT2G45050
|
GATA2, GATA transcription factor 2 |
0.208 |
-0.724 |
| 44 |
261940_at |
AT1G22520
|
unknown |
0.203 |
0.147 |
| 45 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
-1.410 |
-0.930 |
| 46 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
-1.408 |
-0.596 |
| 47 |
266244_at |
AT2G27740
|
unknown |
-1.400 |
-0.113 |
| 48 |
257757_at |
AT3G18660
|
GUX1, glucuronic acid substitution of xylan 1, PGSIP1, plant glycogenin-like starch initiation protein 1 |
-1.364 |
-0.193 |
| 49 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-1.338 |
-0.305 |
| 50 |
263028_at |
AT1G24030
|
[Protein kinase superfamily protein] |
-1.273 |
-0.502 |
| 51 |
264559_at |
AT1G09610
|
GXM3, glucuronoxylan methyltransferase 3 |
-1.224 |
-1.132 |
| 52 |
251723_at |
AT3G56230
|
[BTB/POZ domain-containing protein] |
-1.177 |
0.088 |
| 53 |
265463_at |
AT2G37090
|
IRX9, IRREGULAR XYLEM 9 |
-1.121 |
0.111 |
| 54 |
247030_at |
AT5G67210
|
IRX15-L, IRX15-LIKE |
-1.116 |
0.730 |
| 55 |
249070_at |
AT5G44030
|
CESA4, cellulose synthase A4, IRX5, IRREGULAR XYLEM 5, NWS2 |
-1.110 |
-0.956 |
| 56 |
252944_at |
AT4G39320
|
[microtubule-associated protein-related] |
-1.069 |
-1.147 |
| 57 |
246512_at |
AT5G15630
|
COBL4, COBRA-LIKE4, IRX6, IRREGULAR XYLEM 6 |
-1.044 |
-1.032 |
| 58 |
259606_at |
AT1G27920
|
MAP65-8, microtubule-associated protein 65-8 |
-1.006 |
-1.096 |
| 59 |
254277_at |
AT4G22680
|
AtMYB85, myb domain protein 85, MYB85, myb domain protein 85 |
-0.991 |
-0.319 |
| 60 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
-0.986 |
-0.366 |
| 61 |
246425_at |
AT5G17420
|
ATCESA7, CESA7, CELLULOSE SYNTHASE CATALYTIC SUBUNIT 7, IRX3, IRREGULAR XYLEM 3, MUR10, MURUS 10 |
-0.982 |
0.485 |
| 62 |
262438_at |
AT1G47410
|
unknown |
-0.980 |
-0.290 |
| 63 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.976 |
-0.405 |
| 64 |
256367_at |
AT1G66810
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
-0.952 |
-0.721 |
| 65 |
250933_at |
AT5G03170
|
ATFLA11, ARABIDOPSIS FASCICLIN-LIKE ARABINOGALACTAN-PROTEIN 11, FLA11, FASCICLIN-like arabinogalactan-protein 11 |
-0.931 |
-0.454 |
| 66 |
247590_at |
AT5G60720
|
unknown |
-0.931 |
0.302 |
| 67 |
253877_at |
AT4G27435
|
unknown |
-0.929 |
-0.041 |
| 68 |
255976_at |
AT1G22010
|
unknown |
-0.905 |
-0.328 |
| 69 |
258267_at |
AT3G15870
|
[Fatty acid desaturase family protein] |
-0.894 |
-0.625 |
| 70 |
250120_at |
AT5G16490
|
RIC4, ROP-interactive CRIB motif-containing protein 4 |
-0.850 |
-0.391 |
| 71 |
254618_at |
AT4G18780
|
ATCESA8, CELLULOSE SYNTHASE 8, CESA8, CELLULOSE SYNTHASE 8, IRX1, IRREGULAR XYLEM 1, LEW2, LEAF WILTING 2 |
-0.840 |
0.035 |
| 72 |
247648_at |
AT5G60020
|
ATLAC17, LAC17, laccase 17 |
-0.823 |
0.493 |
| 73 |
252253_at |
AT3G49300
|
[proline-rich family protein] |
-0.798 |
-0.398 |
| 74 |
267037_at |
AT2G38320
|
TBL34, TRICHOME BIREFRINGENCE-LIKE 34 |
-0.792 |
-0.339 |
| 75 |
266221_at |
AT2G28760
|
UXS6, UDP-XYL synthase 6 |
-0.779 |
0.492 |
| 76 |
262795_at |
AT1G13130
|
[Cellulase (glycosyl hydrolase family 5) protein] |
-0.774 |
0.047 |
| 77 |
255923_at |
AT1G22180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.758 |
-0.705 |
| 78 |
266196_at |
AT2G39110
|
[Protein kinase superfamily protein] |
-0.752 |
-0.994 |
| 79 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.750 |
-1.076 |
| 80 |
257502_at |
AT1G78110
|
unknown |
-0.748 |
-0.125 |
| 81 |
248750_at |
AT5G47530
|
[Auxin-responsive family protein] |
-0.742 |
0.307 |
| 82 |
253798_at |
AT4G28500
|
ANAC073, NAC domain containing protein 73, NAC073, NAC domain containing protein 73, SND2, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 2 |
-0.739 |
-0.248 |
| 83 |
251303_at |
AT3G61910
|
ANAC066, NAC domain protein 66, NAC066, NAC domain protein 66, NST2, NAC SECONDARY WALL THICKENING PROMOTING FACTOR2 |
-0.719 |
0.349 |
| 84 |
245174_at |
AT2G47500
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.705 |
0.211 |
| 85 |
262569_at |
AT1G15180
|
[MATE efflux family protein] |
-0.699 |
-0.006 |
| 86 |
248977_at |
AT5G45020
|
[Glutathione S-transferase family protein] |
-0.673 |
-0.097 |
| 87 |
261106_at |
AT1G62990
|
IXR11, KNAT7, KNOTTED-like homeobox of Arabidopsis thaliana 7 |
-0.665 |
-0.827 |