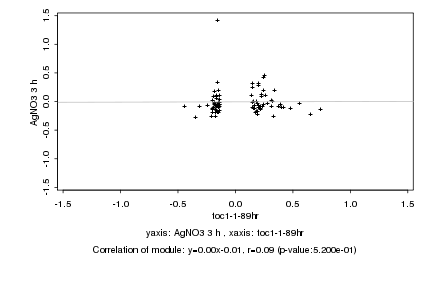
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
toc1-1-89hr |
Log2 signal ratio
AgNO3 3 h |
| 1 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
0.739 |
-0.137 |
| 2 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
0.652 |
-0.211 |
| 3 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
0.551 |
-0.025 |
| 4 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
0.472 |
-0.111 |
| 5 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.416 |
-0.088 |
| 6 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.393 |
-0.096 |
| 7 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.387 |
-0.035 |
| 8 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
0.370 |
-0.075 |
| 9 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
0.336 |
0.195 |
| 10 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
0.331 |
-0.245 |
| 11 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.318 |
0.006 |
| 12 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.306 |
0.030 |
| 13 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.306 |
-0.086 |
| 14 |
266363_at |
AT2G41250
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.271 |
-0.030 |
| 15 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
0.259 |
0.112 |
| 16 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
0.249 |
0.470 |
| 17 |
245702_at |
AT5G04220
|
ATSYTC, NTMC2T1.3, NTMC2TYPE1.3, SYT3, synaptotagmin 3, SYTC |
0.242 |
0.207 |
| 18 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.241 |
0.430 |
| 19 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.236 |
-0.048 |
| 20 |
252917_at |
AT4G38960
|
BBX19, B-box domain protein 19 |
0.232 |
-0.074 |
| 21 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
0.221 |
0.140 |
| 22 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
0.220 |
0.088 |
| 23 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.215 |
-0.110 |
| 24 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.214 |
-0.124 |
| 25 |
259275_at |
AT3G01060
|
unknown |
0.213 |
-0.079 |
| 26 |
248205_at |
AT5G54300
|
unknown |
0.197 |
0.287 |
| 27 |
246095_at |
AT5G19310
|
CHR23, chromatin remodeling 23 |
0.196 |
-0.060 |
| 28 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.195 |
0.315 |
| 29 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
0.192 |
-0.089 |
| 30 |
260425_at |
AT1G72440
|
EDA25, embryo sac development arrest 25, SWA2, SLOW WALKER2 |
0.191 |
-0.164 |
| 31 |
255958_at |
AT1G22150
|
SULTR1;3, sulfate transporter 1;3 |
0.190 |
-0.028 |
| 32 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
0.184 |
-0.224 |
| 33 |
259686_at |
AT1G63100
|
[GRAS family transcription factor] |
0.183 |
0.017 |
| 34 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
0.175 |
-0.167 |
| 35 |
262854_at |
AT1G20870
|
[HSP20-like chaperones superfamily protein] |
0.173 |
-0.188 |
| 36 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.165 |
-0.058 |
| 37 |
247025_at |
AT5G67030
|
ABA1, ABA DEFICIENT 1, ATABA1, ARABIDOPSIS THALIANA ABA DEFICIENT 1, ATZEP, ARABIDOPSIS THALIANA ZEAXANTHIN EPOXIDASE, IBS3, IMPAIRED IN BABA-INDUCED STERILITY 3, LOS6, LOW EXPRESSION OF OSMOTIC STRESS-RESPONSIVE GENES 6, NPQ2, NON-PHOTOCHEMICAL QUENCHING 2, ZEP, ZEAXANTHIN EPOXIDASE |
0.163 |
-0.111 |
| 38 |
255675_at |
AT4G00480
|
ATMYC1, myc1 |
0.157 |
-0.120 |
| 39 |
251005_at |
AT5G02590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.152 |
0.015 |
| 40 |
267637_at |
AT2G42190
|
unknown |
0.147 |
-0.099 |
| 41 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
0.146 |
0.320 |
| 42 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
0.144 |
-0.007 |
| 43 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
0.140 |
0.257 |
| 44 |
249963_at |
AT5G19000
|
ATBPM1, BTB-POZ AND MATH DOMAIN 1, BPM1, BTB-POZ and MATH domain 1 |
0.137 |
0.106 |
| 45 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-0.444 |
-0.085 |
| 46 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.348 |
-0.264 |
| 47 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.320 |
-0.079 |
| 48 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
-0.251 |
-0.062 |
| 49 |
250664_at |
AT5G07080
|
[HXXXD-type acyl-transferase family protein] |
-0.214 |
-0.253 |
| 50 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.208 |
-0.112 |
| 51 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.206 |
-0.185 |
| 52 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
-0.204 |
0.032 |
| 53 |
245568_at |
AT4G14650
|
unknown |
-0.202 |
-0.123 |
| 54 |
253243_at |
AT4G34560
|
unknown |
-0.199 |
0.103 |
| 55 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
-0.193 |
-0.102 |
| 56 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
-0.186 |
-0.024 |
| 57 |
248951_at |
AT5G45550
|
MOB1-like, MOB1-like |
-0.186 |
-0.044 |
| 58 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
-0.185 |
0.188 |
| 59 |
266613_at |
AT2G14900
|
[Gibberellin-regulated family protein] |
-0.184 |
-0.009 |
| 60 |
265131_at |
AT1G23760
|
JP630, PG3, POLYGALACTURONASE 3 |
-0.178 |
-0.125 |
| 61 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.177 |
-0.176 |
| 62 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
-0.176 |
-0.048 |
| 63 |
264909_at |
AT2G17300
|
unknown |
-0.174 |
-0.252 |
| 64 |
263438_at |
AT2G28660
|
[Chloroplast-targeted copper chaperone protein] |
-0.173 |
0.056 |
| 65 |
253581_at |
AT4G30660
|
[Low temperature and salt responsive protein family] |
-0.172 |
-0.156 |
| 66 |
252866_at |
AT4G39840
|
unknown |
-0.170 |
0.110 |
| 67 |
246378_at |
AT1G57620
|
CYB, CYTOPLASMIC BODIES |
-0.168 |
-0.074 |
| 68 |
246791_at |
AT5G27280
|
[Zim17-type zinc finger protein] |
-0.167 |
0.091 |
| 69 |
245728_at |
AT1G73340
|
[Cytochrome P450 superfamily protein] |
-0.164 |
-0.189 |
| 70 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
-0.163 |
-0.066 |
| 71 |
262219_at |
AT1G74750
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.161 |
0.333 |
| 72 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
-0.158 |
1.433 |
| 73 |
255823_at |
AT2G40470
|
ASL11, ASYMMETRIC LEAVES2-LIKE 11, LBD15, LOB domain-containing protein 15 |
-0.157 |
-0.041 |
| 74 |
254108_at |
AT4G25230
|
RIN2, RPM1 interacting protein 2 |
-0.155 |
0.202 |
| 75 |
256916_at |
AT3G24050
|
GATA1, GATA transcription factor 1 |
-0.154 |
-0.001 |
| 76 |
251863_at |
AT3G54870
|
ARK1, ARMADILLO REPEAT-CONTAINING KINESIN 1, AtKINUc, Arabidopsis thaliana KINESIN Ungrouped clade, gene A, CAE1, CA-ROP2 ENHANCER 1, MRH2, MORPHOGENESIS OF ROOT HAIR 2 |
-0.154 |
-0.184 |
| 77 |
247068_at |
AT5G66800
|
unknown |
-0.153 |
-0.043 |
| 78 |
250933_at |
AT5G03170
|
ATFLA11, ARABIDOPSIS FASCICLIN-LIKE ARABINOGALACTAN-PROTEIN 11, FLA11, FASCICLIN-like arabinogalactan-protein 11 |
-0.152 |
-0.084 |
| 79 |
257832_at |
AT3G26740
|
CCL, CCR-like |
-0.151 |
-0.067 |
| 80 |
253205_at |
AT4G34490
|
ATCAP1, cyclase associated protein 1, CAP 1, CAP1, cyclase associated protein 1 |
-0.150 |
-0.104 |
| 81 |
264208_at |
AT1G22760
|
PAB3, poly(A) binding protein 3 |
-0.146 |
0.107 |
| 82 |
260506_at |
AT1G47210
|
CYCA3;2, cyclin-dependent protein kinase 3;2 |
-0.146 |
-0.052 |
| 83 |
259915_at |
AT1G72790
|
[hydroxyproline-rich glycoprotein family protein] |
-0.144 |
-0.051 |
| 84 |
260434_at |
AT1G68330
|
unknown |
-0.144 |
0.042 |
| 85 |
262297_at |
AT1G27600
|
I9H, IRREGULAR XYLEM 9 Homolog, IRX9-L, IRREGULAR XYLEM 9-LIKE |
-0.143 |
-0.003 |
| 86 |
251688_at |
AT3G56480
|
[myosin heavy chain-related] |
-0.142 |
-0.152 |
| 87 |
249444_at |
AT5G39370
|
[Curculin-like (mannose-binding) lectin family protein] |
-0.140 |
-0.075 |