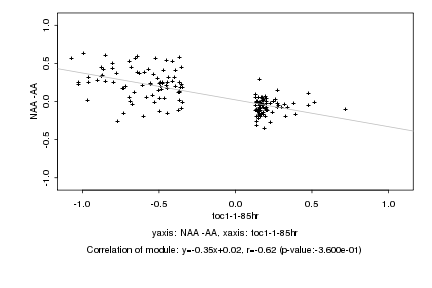
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
toc1-1-85hr |
Log2 signal ratio
NAA -AA |
| 1 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
0.718 |
-0.094 |
| 2 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.516 |
-0.004 |
| 3 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.476 |
0.106 |
| 4 |
265892_at |
AT2G15020
|
unknown |
0.474 |
-0.046 |
| 5 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
0.391 |
-0.166 |
| 6 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.373 |
-0.015 |
| 7 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
0.336 |
-0.068 |
| 8 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.322 |
-0.190 |
| 9 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.318 |
-0.032 |
| 10 |
259927_at |
AT1G75100
|
JAC1, J-domain protein required for chloroplast accumulation response 1 |
0.298 |
-0.076 |
| 11 |
264694_at |
AT1G70250
|
[receptor serine/threonine kinase, putative] |
0.275 |
-0.050 |
| 12 |
245936_at |
AT5G19850
|
[alpha/beta-Hydrolases superfamily protein] |
0.271 |
-0.077 |
| 13 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.271 |
0.145 |
| 14 |
263380_at |
AT2G40200
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.269 |
-0.022 |
| 15 |
261708_at |
AT1G32740
|
[SBP (S-ribonuclease binding protein) family protein] |
0.260 |
0.028 |
| 16 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
0.247 |
0.005 |
| 17 |
260996_at |
AT1G12210
|
RFL1, RPS5-like 1 |
0.243 |
0.013 |
| 18 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.243 |
0.007 |
| 19 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
0.238 |
-0.143 |
| 20 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.226 |
-0.265 |
| 21 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.223 |
-0.018 |
| 22 |
253425_at |
AT4G32190
|
[Myosin heavy chain-related protein] |
0.207 |
-0.117 |
| 23 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
0.202 |
-0.114 |
| 24 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
0.201 |
0.051 |
| 25 |
249112_at |
AT5G43780
|
APS4 |
0.200 |
0.003 |
| 26 |
263122_at |
AT1G78510
|
AtSPS1, SPS1, solanesyl diphosphate synthase 1 |
0.199 |
-0.074 |
| 27 |
256483_at |
AT1G31410
|
ENF2, ENLARGED FIL EXPRESSION DOMAIN 2 |
0.199 |
-0.104 |
| 28 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
0.197 |
-0.021 |
| 29 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
0.194 |
-0.195 |
| 30 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
0.193 |
0.069 |
| 31 |
259065_at |
AT3G07520
|
ATGLR1.4, GLR1.4, glutamate receptor 1.4 |
0.190 |
-0.087 |
| 32 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.184 |
-0.345 |
| 33 |
258726_at |
AT3G11745
|
unknown |
0.183 |
-0.071 |
| 34 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
0.183 |
-0.167 |
| 35 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
0.181 |
0.026 |
| 36 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
0.180 |
0.035 |
| 37 |
259762_at |
AT1G77600
|
[ARM repeat superfamily protein] |
0.178 |
-0.027 |
| 38 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
0.177 |
-0.137 |
| 39 |
253042_at |
AT4G37550
|
[Acetamidase/Formamidase family protein] |
0.174 |
-0.049 |
| 40 |
266862_at |
AT2G26950
|
AtMYB104, myb domain protein 104, MYB104, myb domain protein 104 |
0.173 |
-0.043 |
| 41 |
249963_at |
AT5G19000
|
ATBPM1, BTB-POZ AND MATH DOMAIN 1, BPM1, BTB-POZ and MATH domain 1 |
0.171 |
0.042 |
| 42 |
253814_at |
AT4G28290
|
unknown |
0.171 |
0.024 |
| 43 |
258677_at |
AT3G08730
|
ATPK1, ARABIDOPSIS THALIANA PROTEIN-SERINE KINASE 1, ATPK6, ARABIDOPSIS THALIANA PROTEIN-SERINE KINASE 6, ATS6K1, PK1, protein-serine kinase 1, PK6, ROTEIN-SERINE KINASE 6, S6K1, P70 RIBOSOMAL S6 KINASE |
0.171 |
0.054 |
| 44 |
261089_at |
AT1G07570
|
APK1, APK1A |
0.169 |
-0.024 |
| 45 |
259460_at |
AT1G44000
|
unknown |
0.168 |
-0.149 |
| 46 |
259275_at |
AT3G01060
|
unknown |
0.168 |
-0.159 |
| 47 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.166 |
0.053 |
| 48 |
248189_at |
AT5G54090
|
[DNA mismatch repair protein MutS, type 2] |
0.163 |
-0.189 |
| 49 |
259856_at |
AT1G68440
|
unknown |
0.162 |
-0.006 |
| 50 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.160 |
-0.115 |
| 51 |
260137_at |
AT1G66330
|
[senescence-associated family protein] |
0.160 |
-0.052 |
| 52 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.159 |
-0.086 |
| 53 |
252089_at |
AT3G52110
|
unknown |
0.157 |
-0.069 |
| 54 |
246370_at |
AT1G51920
|
unknown |
0.157 |
0.293 |
| 55 |
256032_at |
AT1G34090
|
[copia-like retrotransposon family, has a 8.8e-131 P-value blast match to GB:BAA78423 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)GB:BAA78423 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)GB:BAA78423 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)gi|4996361|dbj|BAA78423.1| polyprotein (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.155 |
-0.043 |
| 56 |
246058_at |
AT5G08430
|
[SWIB/MDM2 domain] |
0.155 |
-0.007 |
| 57 |
250613_at |
AT5G07240
|
IQD24, IQ-domain 24 |
0.152 |
-0.167 |
| 58 |
247560_at |
AT5G61090
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.152 |
-0.157 |
| 59 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
0.152 |
-0.178 |
| 60 |
260633_at |
AT1G62400
|
HT1, high leaf temperature 1 |
0.150 |
-0.110 |
| 61 |
252309_at |
AT3G49340
|
[Cysteine proteinases superfamily protein] |
0.149 |
-0.023 |
| 62 |
249321_at |
AT5G40920
|
[pseudogene] |
0.149 |
-0.102 |
| 63 |
247594_at |
AT5G60800
|
[Heavy metal transport/detoxification superfamily protein ] |
0.149 |
-0.126 |
| 64 |
256425_at |
AT1G33560
|
ADR1, ACTIVATED DISEASE RESISTANCE 1 |
0.148 |
0.056 |
| 65 |
260769_at |
AT1G49010
|
[Duplicated homeodomain-like superfamily protein] |
0.147 |
-0.214 |
| 66 |
254068_at |
AT4G25450
|
ABCB28, ATP-binding cassette B28, ATNAP8, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 8, NAP8, non-intrinsic ABC protein 8 |
0.146 |
-0.024 |
| 67 |
247694_at |
AT5G59750
|
AtRIBA3, RIBA3, homolog of ribA 3 |
0.145 |
-0.113 |
| 68 |
256020_at |
AT1G58290
|
AtHEMA1, Arabidopsis thaliana hemA 1, HEMA1 |
0.142 |
-0.109 |
| 69 |
250480_at |
AT5G10290
|
[leucine-rich repeat transmembrane protein kinase family protein] |
0.140 |
0.008 |
| 70 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.140 |
-0.122 |
| 71 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.136 |
-0.310 |
| 72 |
253131_at |
AT4G35550
|
ATWOX13, HB-4, WOX13, WUSCHEL related homeobox 13 |
0.135 |
-0.010 |
| 73 |
248595_at |
AT5G49230
|
HRB1, HYPERSENSITIVE TO RED AND BLUE |
0.135 |
-0.007 |
| 74 |
264435_at |
AT1G10360
|
ATGSTU18, glutathione S-transferase TAU 18, GST29, GLUTATHIONE S-TRANSFERASE 29, GSTU18, glutathione S-transferase TAU 18 |
0.135 |
-0.252 |
| 75 |
248988_at |
AT5G45190
|
[Cyclin family protein] |
0.135 |
0.008 |
| 76 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
0.133 |
-0.193 |
| 77 |
264496_at |
AT1G27285
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:AAC02666 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
0.132 |
-0.100 |
| 78 |
267445_at |
AT2G33680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.131 |
-0.048 |
| 79 |
253434_at |
AT4G32500
|
AKT5, K+ transporter 5, KT5, K+ transporter 5 |
0.131 |
-0.107 |
| 80 |
246983_at |
AT5G67200
|
[Leucine-rich repeat protein kinase family protein] |
0.130 |
0.096 |
| 81 |
246840_at |
AT5G26610
|
[D111/G-patch domain-containing protein] |
0.130 |
0.056 |
| 82 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-1.076 |
0.567 |
| 83 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-1.031 |
0.232 |
| 84 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-1.026 |
0.253 |
| 85 |
246652_at |
AT5G35190
|
EXT13, extensin 13 |
-0.994 |
0.635 |
| 86 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.970 |
0.026 |
| 87 |
250778_at |
AT5G05500
|
MOP10 |
-0.966 |
0.316 |
| 88 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.963 |
0.255 |
| 89 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.903 |
0.285 |
| 90 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.880 |
0.334 |
| 91 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.878 |
0.447 |
| 92 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.873 |
0.352 |
| 93 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
-0.868 |
0.433 |
| 94 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
-0.852 |
0.274 |
| 95 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.850 |
0.610 |
| 96 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.805 |
0.507 |
| 97 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.804 |
0.440 |
| 98 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.803 |
0.258 |
| 99 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.783 |
0.378 |
| 100 |
259276_at |
AT3G01190
|
[Peroxidase superfamily protein] |
-0.771 |
-0.250 |
| 101 |
245172_at |
AT2G47540
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.739 |
0.181 |
| 102 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
-0.738 |
-0.147 |
| 103 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.735 |
0.182 |
| 104 |
250059_at |
AT5G17820
|
[Peroxidase superfamily protein] |
-0.719 |
0.199 |
| 105 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
-0.698 |
0.062 |
| 106 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
-0.693 |
0.529 |
| 107 |
262658_at |
AT1G14220
|
[Ribonuclease T2 family protein] |
-0.686 |
0.013 |
| 108 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.683 |
0.459 |
| 109 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
-0.675 |
-0.036 |
| 110 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.660 |
0.122 |
| 111 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.659 |
0.566 |
| 112 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.645 |
0.591 |
| 113 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.644 |
0.384 |
| 114 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.629 |
0.377 |
| 115 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.611 |
0.217 |
| 116 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
-0.604 |
-0.188 |
| 117 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-0.599 |
0.383 |
| 118 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.586 |
0.053 |
| 119 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.569 |
0.422 |
| 120 |
251405_at |
AT3G60330
|
AHA7, H(+)-ATPase 7, HA7, H(+)-ATPase 7 |
-0.561 |
0.246 |
| 121 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.556 |
0.230 |
| 122 |
249375_at |
AT5G40730
|
AGP24, arabinogalactan protein 24, ATAGP24, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEIN 24 |
-0.543 |
0.080 |
| 123 |
247094_at |
AT5G66280
|
GMD1, GDP-D-mannose 4,6-dehydratase 1 |
-0.536 |
0.364 |
| 124 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
-0.535 |
-0.007 |
| 125 |
261691_at |
AT1G50060
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.523 |
0.573 |
| 126 |
253613_at |
AT4G30320
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.512 |
0.306 |
| 127 |
261999_at |
AT1G33800
|
AtGXMT1, GXM1, glucuronoxylan methyltransferase 1, GXMT1, glucuronoxylan methyltransferase 1 |
-0.504 |
0.145 |
| 128 |
264577_at |
AT1G05260
|
RCI3, RARE COLD INDUCIBLE GENE 3, RCI3A |
-0.502 |
0.043 |
| 129 |
267409_at |
AT2G34910
|
unknown |
-0.502 |
0.245 |
| 130 |
247965_at |
AT5G56540
|
AGP14, arabinogalactan protein 14, ATAGP14 |
-0.499 |
-0.118 |
| 131 |
249009_at |
AT5G44610
|
MAP18, microtubule-associated protein 18, PCAP2, PLASMA MEMBRANE ASSOCIATED CA2+-BINDING PROTEIN-2 |
-0.495 |
0.256 |
| 132 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.490 |
0.208 |
| 133 |
250469_at |
AT5G10130
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.486 |
0.163 |
| 134 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
-0.482 |
0.250 |
| 135 |
262349_at |
AT2G48130
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.481 |
0.241 |
| 136 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
-0.472 |
0.413 |
| 137 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
-0.464 |
0.044 |
| 138 |
249756_at |
AT5G24313
|
unknown |
-0.457 |
0.540 |
| 139 |
250683_x_at |
AT5G06640
|
EXT10, extensin 10 |
-0.452 |
0.212 |
| 140 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.450 |
0.252 |
| 141 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.448 |
-0.146 |
| 142 |
251843_x_at |
AT3G54590
|
ATHRGP1, hydroxyproline-rich glycoprotein, HRGP1, hydroxyproline-rich glycoprotein |
-0.446 |
0.181 |
| 143 |
259328_at |
AT3G16440
|
ATMLP-300B, myrosinase-binding protein-like protein-300B, MEE36, maternal effect embryo arrest 36, MLP-300B, myrosinase-binding protein-like protein-300B |
-0.442 |
0.327 |
| 144 |
262045_at |
AT1G80240
|
DGR1, DUF642 L-GalL responsive gene 1 |
-0.417 |
0.537 |
| 145 |
262317_at |
AT2G48140
|
EDA4, embryo sac development arrest 4 |
-0.412 |
0.275 |
| 146 |
267191_at |
AT2G44110
|
ATMLO15, MILDEW RESISTANCE LOCUS O 15, MLO15, MILDEW RESISTANCE LOCUS O 15 |
-0.400 |
0.325 |
| 147 |
250224_at |
AT5G14150
|
unknown |
-0.397 |
0.208 |
| 148 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
-0.396 |
0.417 |
| 149 |
259964_at |
AT1G53680
|
ATGSTU28, glutathione S-transferase TAU 28, GSTU28, glutathione S-transferase TAU 28 |
-0.377 |
-0.105 |
| 150 |
265049_at |
AT1G52060
|
[Mannose-binding lectin superfamily protein] |
-0.375 |
0.131 |
| 151 |
266591_at |
AT2G46225
|
ABIL1, ABI-1-like 1 |
-0.372 |
0.025 |
| 152 |
249358_at |
AT5G40510
|
[Sucrase/ferredoxin-like family protein] |
-0.371 |
0.143 |
| 153 |
266356_at |
AT2G32300
|
UCC1, uclacyanin 1 |
-0.370 |
0.127 |
| 154 |
245139_at |
AT2G45430
|
AHL22, AT-hook motif nuclear-localized protein 22 |
-0.370 |
0.256 |
| 155 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
-0.369 |
0.579 |
| 156 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.364 |
0.192 |
| 157 |
257924_at |
AT3G23190
|
[HR-like lesion-inducing protein-related] |
-0.358 |
0.236 |
| 158 |
248626_at |
AT5G48940
|
[Leucine-rich repeat transmembrane protein kinase family protein] |
-0.356 |
0.457 |
| 159 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.353 |
-0.090 |
| 160 |
266920_at |
AT2G45750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.351 |
0.187 |
| 161 |
265052_at |
AT1G51990
|
[O-methyltransferase family protein] |
-0.348 |
-0.006 |