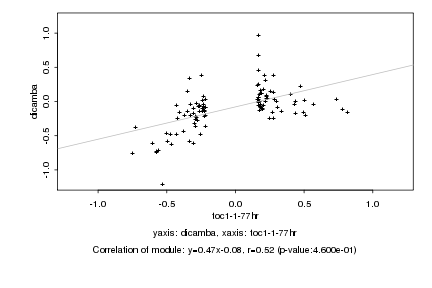
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
toc1-1-77hr |
Log2 signal ratio
dicamba |
| 1 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.809 |
-0.155 |
| 2 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.774 |
-0.107 |
| 3 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.733 |
0.039 |
| 4 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.565 |
-0.029 |
| 5 |
257670_at |
AT3G20340
|
unknown |
0.508 |
-0.190 |
| 6 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.500 |
0.023 |
| 7 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.492 |
-0.154 |
| 8 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.472 |
0.229 |
| 9 |
262452_at |
AT1G11210
|
unknown |
0.437 |
0.006 |
| 10 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.435 |
-0.168 |
| 11 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.424 |
-0.037 |
| 12 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.400 |
0.113 |
| 13 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.329 |
-0.139 |
| 14 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.306 |
-0.075 |
| 15 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
0.292 |
0.009 |
| 16 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
0.279 |
0.042 |
| 17 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.277 |
0.391 |
| 18 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.271 |
0.141 |
| 19 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.270 |
-0.248 |
| 20 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
0.269 |
-0.160 |
| 21 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.251 |
0.159 |
| 22 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.241 |
-0.243 |
| 23 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.230 |
0.056 |
| 24 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.222 |
0.056 |
| 25 |
254948_at |
AT4G11000
|
[Ankyrin repeat family protein] |
0.222 |
0.097 |
| 26 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.219 |
0.087 |
| 27 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
0.217 |
0.007 |
| 28 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.214 |
0.322 |
| 29 |
256430_at |
AT3G11020
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2B, DRE/CRT-binding protein 2B |
0.209 |
0.392 |
| 30 |
245765_at |
AT1G33600
|
[Leucine-rich repeat (LRR) family protein] |
0.202 |
0.185 |
| 31 |
253506_at |
AT4G31980
|
unknown |
0.199 |
-0.057 |
| 32 |
247937_at |
AT5G57110
|
ACA8, autoinhibited Ca2+ -ATPase, isoform 8, AT-ACA8, AUTOINHIBITED CA2+ -ATPASE, ISOFORM 8 |
0.197 |
-0.100 |
| 33 |
261318_at |
AT1G53035
|
unknown |
0.191 |
-0.111 |
| 34 |
264787_at |
AT2G17840
|
ERD7, EARLY-RESPONSIVE TO DEHYDRATION 7 |
0.186 |
0.126 |
| 35 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.185 |
0.127 |
| 36 |
262296_at |
AT1G27630
|
CYCT1;3, cyclin T 1;3 |
0.181 |
-0.061 |
| 37 |
253747_at |
AT4G29050
|
LecRK-V.9, L-type lectin receptor kinase V.9 |
0.179 |
-0.035 |
| 38 |
256061_at |
AT1G07040
|
unknown |
0.179 |
0.123 |
| 39 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.178 |
0.174 |
| 40 |
254437_s_at |
AT3G06433
|
[pseudogene] |
0.178 |
-0.080 |
| 41 |
249977_at |
AT5G18820
|
Cpn60alpha2, chaperonin-60alpha2, EMB3007, embryo defective 3007 |
0.172 |
0.012 |
| 42 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
0.171 |
0.115 |
| 43 |
250412_at |
AT5G11150
|
ATVAMP713, vesicle-associated membrane protein 713, VAMP713, VAMP713, vesicle-associated membrane protein 713 |
0.170 |
-0.128 |
| 44 |
252468_at |
AT3G46970
|
ATPHS2, Arabidopsis thaliana alpha-glucan phosphorylase 2, PHS2, alpha-glucan phosphorylase 2 |
0.166 |
0.035 |
| 45 |
249586_at |
AT5G37840
|
unknown |
0.166 |
0.014 |
| 46 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.166 |
0.970 |
| 47 |
260359_at |
AT1G69210
|
[Uncharacterised protein family UPF0090] |
0.165 |
-0.053 |
| 48 |
259489_at |
AT1G15790
|
unknown |
0.164 |
0.459 |
| 49 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.163 |
0.249 |
| 50 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
0.163 |
0.073 |
| 51 |
248510_at |
AT5G50315
|
[Mutator-like transposase family, has a 5.0e-14 P-value blast match to Q9XE24 /118-277 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
0.162 |
-0.012 |
| 52 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
0.161 |
0.065 |
| 53 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.161 |
0.674 |
| 54 |
251603_at |
AT3G57760
|
[Protein kinase superfamily protein] |
0.160 |
0.239 |
| 55 |
251789_at |
AT3G55450
|
PBL1, PBS1-like 1 |
0.156 |
0.042 |
| 56 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.752 |
-0.747 |
| 57 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.728 |
-0.376 |
| 58 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.607 |
-0.603 |
| 59 |
250778_at |
AT5G05500
|
MOP10 |
-0.582 |
-0.738 |
| 60 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.578 |
-0.718 |
| 61 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.561 |
-0.710 |
| 62 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.533 |
-1.199 |
| 63 |
246652_at |
AT5G35190
|
EXT13, extensin 13 |
-0.509 |
-0.455 |
| 64 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.501 |
-0.579 |
| 65 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
-0.474 |
-0.473 |
| 66 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.466 |
-0.617 |
| 67 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
-0.433 |
-0.480 |
| 68 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.431 |
-0.045 |
| 69 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.428 |
-0.247 |
| 70 |
249375_at |
AT5G40730
|
AGP24, arabinogalactan protein 24, ATAGP24, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEIN 24 |
-0.410 |
-0.154 |
| 71 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.383 |
-0.429 |
| 72 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
-0.376 |
-0.197 |
| 73 |
247965_at |
AT5G56540
|
AGP14, arabinogalactan protein 14, ATAGP14 |
-0.356 |
-0.136 |
| 74 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.355 |
0.147 |
| 75 |
245172_at |
AT2G47540
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.337 |
-0.574 |
| 76 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.336 |
0.337 |
| 77 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.333 |
-0.192 |
| 78 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.328 |
-0.042 |
| 79 |
246949_at |
AT5G25140
|
CYP71B13, cytochrome P450, family 71, subfamily B, polypeptide 13 |
-0.311 |
-0.099 |
| 80 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.311 |
-0.608 |
| 81 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.311 |
-0.164 |
| 82 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
-0.302 |
-0.310 |
| 83 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
-0.298 |
-0.261 |
| 84 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.298 |
-0.360 |
| 85 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.291 |
-0.215 |
| 86 |
264577_at |
AT1G05260
|
RCI3, RARE COLD INDUCIBLE GENE 3, RCI3A |
-0.287 |
-0.017 |
| 87 |
266838_at |
AT2G25980
|
[Mannose-binding lectin superfamily protein] |
-0.286 |
-0.230 |
| 88 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.281 |
-0.277 |
| 89 |
246439_at |
AT5G17600
|
[RING/U-box superfamily protein] |
-0.269 |
-0.061 |
| 90 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.268 |
-0.142 |
| 91 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.266 |
-0.055 |
| 92 |
256097_at |
AT1G13670
|
unknown |
-0.263 |
-0.060 |
| 93 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.261 |
-0.479 |
| 94 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.253 |
0.389 |
| 95 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.246 |
0.029 |
| 96 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.245 |
-0.099 |
| 97 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.245 |
-0.140 |
| 98 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.237 |
-0.063 |
| 99 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.237 |
0.088 |
| 100 |
254037_at |
AT4G25760
|
ATGDU2, glutamine dumper 2, GDU2, glutamine dumper 2 |
-0.236 |
-0.033 |
| 101 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.235 |
-0.130 |
| 102 |
266391_at |
AT2G41290
|
SSL2, strictosidine synthase-like 2 |
-0.232 |
-0.121 |
| 103 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.227 |
-0.214 |
| 104 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
-0.226 |
-0.135 |
| 105 |
248080_at |
AT5G55380
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.225 |
-0.204 |
| 106 |
258241_at |
AT3G27650
|
LBD25, LOB domain-containing protein 25 |
-0.222 |
-0.085 |
| 107 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-0.221 |
-0.361 |
| 108 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.221 |
-0.074 |
| 109 |
247713_at |
AT5G59330
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.220 |
0.030 |